The Galaxy Docker Image is an easy distributable full-fledged Galaxy installation, that can be used for testing, teaching and presenting new tools and features.
One of the main goals is to make the access to entire tool suites as easy as possible. Usually, this includes the setup of a public available web-service that needs to be maintained, or that the Tool-user needs to either setup a Galaxy Server by its own or to have Admin access to a local Galaxy server. With docker, tool developers can create their own Image with all dependencies and the user only needs to run it within docker.
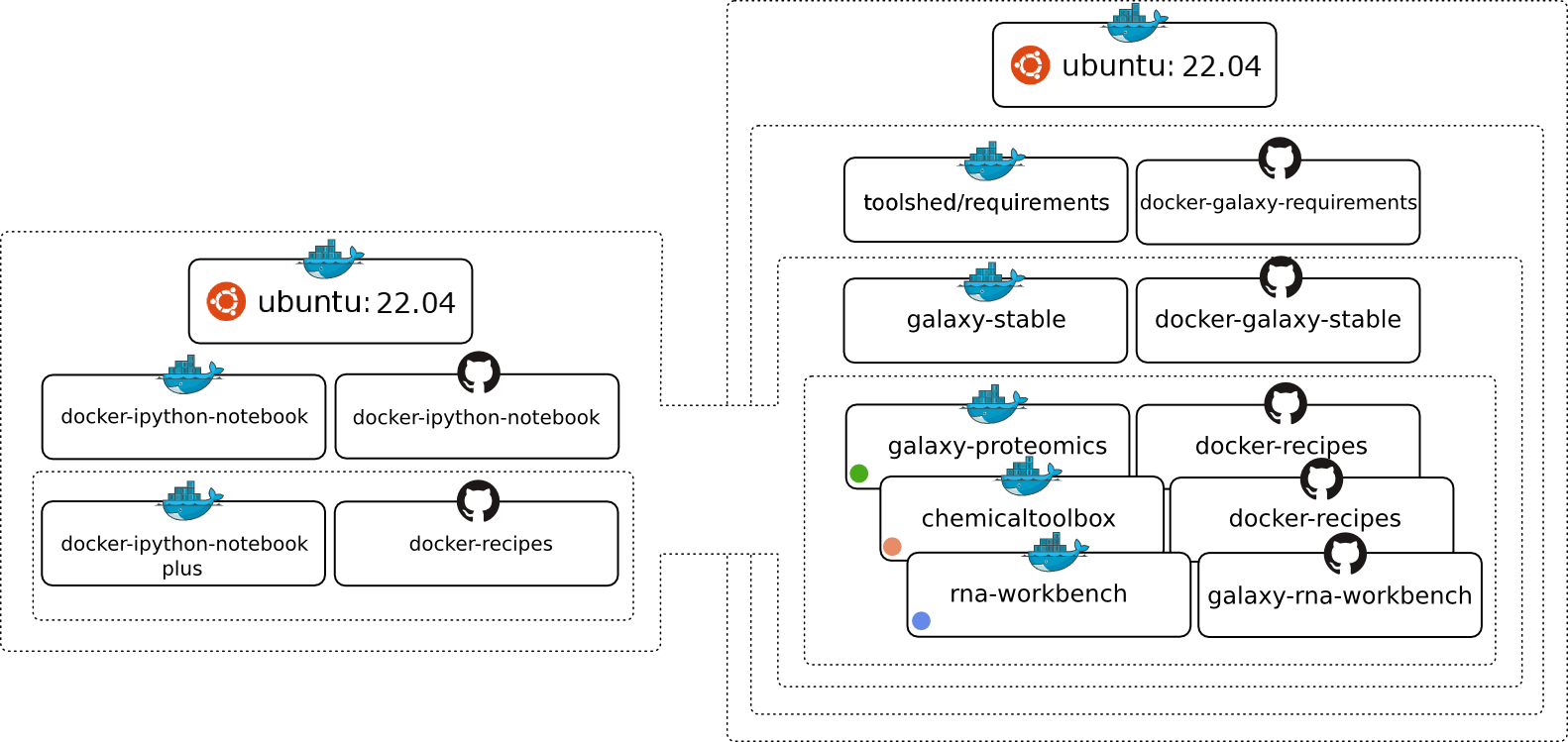
The Image is based on Ubuntu 14.04 LTS and all recommended Galaxy requirements are installed. The following chart should illustrate the Docker image hierarchy we have build to make is as easy as possible to build on different layers of our stack and create many exciting Galaxy flavors.
ℹ️ 19.01 was the last release which is based on ubuntu:14.04 and
PostgreSQL 9.3. In 19.05 we have migrate to ubuntu:18.04 and PostgreSQL version 11.5. Furthermore, we no longer
support old Galaxy tool dependencies. These are deprecated since a few years and we think it's time to remove support
for this in the default installation. You can install all needed packages manually to enable support again, though.
In short, with 19.05:
- Galaxy tool dependencies are be supported by default
ubuntu:18.04is the new base image- a new PostgreSQL version means you need to migrate any old database to the new version
ℹ️ Since 20.05 the default admin password and apikey (GALAXY_DEFAULT_ADMIN_PASSWORD and GALAXY_DEFAULT_ADMIN_KEY) have changed: the password is now password (instead of admin) and the apikey fakekey (instead of admin).
- Usage
- Upgrading images
- Enabling Interactive Environments in Galaxy
- Using passive mode FTP or SFTP
- Using Parent docker
- Galaxy Report Webapp
- Galaxy's config settings
- Configuring Galaxy's behind a proxy
- On-demand reference data with CVMFS
- Personalize your Galaxy
- Deactivating services
- Restarting Galaxy
- Advanced Logging
- Running on an external cluster (DRM)
- Enable Galaxy to use BioContainers (Docker)
- Magic Environment variables
- HTTPS Support
- Lite Mode
- Extending the Docker Image
- Integrating non-Tool Shed tools into the container
- Users & Passwords
- Development
- Requirements
- History
- Support & Bug Reports
Usage [toc]
This chapter explains how to launch the container manually.
At first you need to install docker. Please follow the very good instructions from the Docker project.
After the successful installation, all you need to do is:
docker run -d -p 8080:80 -p 8021:21 -p 8022:22 bgruening/galaxy-stableI will shortly explain the meaning of all the parameters. For a more detailed description please consult the docker manual, it's really worth reading.
Let's start:
-
docker runwill run the Image/Container for you.In case you do not have the Container stored locally, docker will download it for you.
-
-p 8080:80will make the port 80 (inside of the container) available on port 8080 on your host. Same holds for port 8021 and 8022, that can be used to transfer data via the FTP or SFTP protocol, respectively.Inside the container a nginx Webserver is running on port 80 and that port can be bound to a local port on your host computer. With this parameter you can access your Galaxy instance via
http://localhost:8080immediately after executing the command above. If you work with the Docker Toolbox on Mac or Windows, you need to connect to the machine generated by 'Docker Quickstart'. You get its IP address fromdocker-machine lsor from the first line in the terminal, e.g.:docker is configured to use the default machine with IP 192.168.99.100. -
bgruening/galaxy-stableis the Image/Container name, that directs docker to the correct path in the docker index. -
-dwill start the docker container in daemon mode.
For an interactive session, you can execute:
docker run -i -t -p 8080:80 \
bgruening/galaxy-stable \
/bin/bashand run the startup script by yourself, to start PostgreSQL, nginx and Galaxy.
Docker images are "read-only", all your changes inside one session will be lost after restart. This mode is useful to present Galaxy to your colleagues or to run workshops with it. To install Tool Shed repositories or to save your data you need to export the calculated data to the host computer.
Fortunately, this is as easy as:
docker run -d -p 8080:80 \
-v /home/user/galaxy_storage/:/export/ \
bgruening/galaxy-stableWith the additional -v /home/user/galaxy_storage/:/export/ parameter, Docker will mount the local folder /home/user/galaxy_storage into the Container under /export/. A startup.sh script, that is usually starting nginx, PostgreSQL and Galaxy, will recognize the export directory with one of the following outcomes:
- In case of an empty
/export/directory, it will move the PostgreSQL database, the Galaxy database directory, Shed Tools and Tool Dependencies and various config scripts to /export/ and symlink back to the original location. - In case of a non-empty
/export/, for example if you continue a previous session within the same folder, nothing will be moved, but the symlinks will be created.
This enables you to have different export folders for different sessions - means real separation of your different projects.
You can also collect and store /export/ data of Galaxy instances in a dedicated docker Data volume Container created by:
docker create -v /export \
--name galaxy-store \
bgruening/galaxy-stable \
/bin/trueTo mount this data volume in a Galaxy container, use the --volumes-from parameter:
docker run -d -p 8080:80 \
--volumes-from galaxy-store \
bgruening/galaxy-stableThis also allows for data separation, but keeps everything encapsulated within the docker engine (e.g. on OS X within your $HOME/.docker folder - easy to backup, archive and restore. This approach, albeit at the expense of disk space, avoids the problems with permissions reported for data export on non-Linux hosts.
Upgrading images [toc]
We will release a new version of this image concurrent with every new Galaxy release. For upgrading an image to a new version we have assembled a few hints for you. Please, take in account that upgrading may vary depending on your Galaxy installation, and the changes in new versions. Use this example carefully!
- Create a test instance with only the database and configuration files. This will allow testing to ensure that things run but won't require copying all of the data.
- New unmodified configuration files are always stored in a hidden directory called
.distribution_config. Use this folder to diff your configurations with the new configuration files shipped with Galaxy. This prevents needing to go through the change log files to find out which new files were added or which new features you can activate.
Here are 2 suggested upgrade methods, a quick one, and a safer one.
This method involves less data copying, which makes the process quicker, but makes it impossible to downgrade in case of problems.
If you are upgrading from <19.05 to >=19.05, you need to migrate the PostgreSQL database, have a look at PostgreSQL migration.
- Stop the old Galaxy container
docker stop <old_container_name>
docker pull bgruening/galaxy-stable- Run the container with the updated image
docker run -p 8080:80 -v /data/galaxy-data:/export --name <new_container_name> bgruening/galaxy-stable- Use diff to find changes in the config files (only if you changed any config file).
cd /data/galaxy-data/.distribution_config
for f in *; do echo $f; diff $f ../galaxy-central/config/$f; read; done- Upgrade the database schema
docker exec -it <new_container_name> bash
supervisorctl stop galaxy:
sh manage_db.sh upgrade
exit- Restart Galaxy
docker exec -it <new_container_name> supervisorctl start galaxy:(Alternatively, restart the whole container)
With this method, you keep a backup in case you decide to downgrade, but requires some potentially long data copying.
- Note that copying database and datasets can be expensive if you have many GB of data.
- If you are upgrading from <19.05 to >=19.05, you need to migrate the PostgreSQL database, have a look at PostgreSQL migration.
- Download newer version of the Galaxy image
$ sudo docker pull bgruening/galaxy-stable
- Stop and rename the current galaxy container
$ sudo docker stop galaxy-instance
$ sudo docker rename galaxy-instance galaxy-instance-old
- Rename the data directory (the one that is mounted to /export in the docker)
$ sudo mv /data/galaxy-data /data/galaxy-data-old
- Run a new Galaxy container using newer image and wait while Galaxy generates the default content for /export
$ sudo docker run -p 8080:80 -v /data/galaxy-data:/export --name galaxy-instance bgruening/galaxy-stable
- Stop the Galaxy container
$ sudo docker stop galaxy-instance
- Replace the content of the postgres database by the old db data
$ sudo rm -r /data/galaxy-data/postgresql/
$ sudo rsync -var /data/galaxy-data-old/postgresql/ /data/galaxy-data/postgresql/
- Use diff to find changes in the config files (only if you changed any config file).
$ cd /data/galaxy-data/.distribution_config
$ for f in *; do echo $f; diff $f ../../galaxy-data-old/galaxy-central/config/$f; read; done
- Copy all the users' datasets to the new instance
$ sudo rsync -var /data/galaxy-data-old/galaxy-central/database/files/* /data/galaxy-data/galaxy-central/da
tabase/files/
- Copy all the installed tools
$ sudo rsync -var /data/galaxy-data-old/tool_deps/* /data/galaxy-data/tool_deps/
$ sudo rsync -var /data/galaxy-data-old/shed_tools/* /data/galaxy-data/shed_tools/
- Copy the welcome page and all its files.
$ sudo rsync -var /data/galaxy-data-old/welcome* /data/galaxy-data/
- Create an auxiliary docker in interactive mode and upgrade the database.
$ sudo docker run -it --rm -v /data/galaxy-data:/export bgruening/galaxy-stable /bin/bash
# Startup all processes
> startup &
#Upgrade the database to the most recent version
> sh manage_db.sh upgrade
#Logout
> exit
- Start the docker and test
$ sudo docker start galaxy-instance
- Clean the old container and image
Postgresql migration [toc]
In the 19.05 version, Postgresql was updated from version 9.3 to version 11.5. If you are upgrading from a version <19.05, you will need to migrate the database. You can do it the following way (based on the "The quick upgrade method" above):
- Stop Galaxy in the old container
docker exec -it <old_container_name> supervisorctl stop galaxy:- Dump the old database
docker exec -it <old_container_name> bash
su postgres
pg_dumpall --clean > /export/postgresql/9.3dump.sql
exit
exit- Update the container (= step 1 of the "The quick upgrade method" above)
docker stop <old_container_name>
docker pull bgruening/galaxy-stable- Run the container with the updated image (= step 2 of the "The quick upgrade method" above)
docker run -p 8080:80 -v /data/galaxy-data:/export --name <new_container_name> bgruening/galaxy-stable- Restore the dump to the new postgres version
Wait for the startup process to finish (Galaxy should be accessible)
docker exec -it <new_container_name> bash
supervisorctl stop galaxy:
su postgres
psql -f /export/postgresql/9.3dump.sql postgres
exit
exit- Use diff to find changes in the config files (only if you changed any config file). (= step 3 of the "The quick upgrade method" above)
cd /data/galaxy-data/.distribution_config
for f in *; do echo $f; diff $f ../galaxy-central/config/$f; read; done- Upgrade the database schema (= step 4 of the "The quick upgrade method" above)
docker exec -it <new_container_name> bash
supervisorctl stop galaxy:
sh manage_db.sh upgrade
exit- Restart Galaxy (= step 5 of the "The quick upgrade method" above)
docker exec -it <new_container_name> supervisorctl start galaxy:(Alternatively, restart the whole container)
- Clean old files
If you are very sure that everything went well, you can delete /export/postgresql/9.3dump.sql and /export/postgresql/9.3/ to save some space.
Enabling Interactive Environments in Galaxy [toc]
Interactive Environments (IE) are sophisticated ways to extend Galaxy with powerful services, like Jupyter, in a secure and reproducible way.
For this we need to be able to launch Docker containers inside our Galaxy Docker container. At least docker 1.3 is needed on the host system.
docker run -d -p 8080:80 -p 8021:21 -p 8800:8800 \
--privileged=true \
-v /home/user/galaxy_storage/:/export/ \
bgruening/galaxy-stableThe port 8800 is the proxy port that is used to handle Interactive Environments. --privileged is needed to start docker containers inside docker. If your IE does not open, please make sure you open your Galaxy instance with your hostname or a FQDN, but not with localhost or 127.0.0.1.
Using passive mode FTP or SFTP [toc]
By default, FTP servers running inside of docker containers are not accessible via passive mode FTP, due to not being able to expose extra ports. To circumvent this, you can use the --net=host option to allow Docker to directly open ports on the host server:
docker run -d \
--net=host \
-v /home/user/galaxy_storage/:/export/ \
bgruening/galaxy-stableNote that there is no need to specifically bind individual ports (e.g., -p 80:80) if you use --net.
An alternative to FTP and it's shortcomings it to use the SFTP protocol via port 22. Start your Galaxy container with a port binding to 22.
docker run -i -t -p 8080:80 -p 8022:22 \
-v /home/user/galaxy_storage/:/export/ \
bgruening/galaxy-stableAnd use for example Filezilla or the sftp program to transfer data:
sftp -v -P 8022 -o [email protected] localhost <<< $'put <YOUR FILE HERE>'Using Parent docker [toc]
On some linux distributions, Docker-In-Docker can run into issues (such as running out of loopback interfaces). If this is an issue, you can use a 'legacy' mode that use a docker socket for the parent docker installation mounted inside the container. To engage, set the environmental variable DOCKER_PARENT
docker run -p 8080:80 -p 8021:21 -p 8800:8800 \
--privileged=true -e DOCKER_PARENT=True \
-v /var/run/docker.sock:/var/run/docker.sock \
-v /home/user/galaxy_storage/:/export/ \
bgruening/galaxy-stableGalaxy Report Webapp [toc]
For admins wishing to have more information on the status of a galaxy instance, the Galaxy Report Webapp is served on http://localhost:8080/reports. As default this site is password protected with admin:admin. You can change this by providing a reports_htpasswd file in /home/user/galaxy_storage/.
You can disable the Report Webapp entirely by providing the environment variable NONUSE during container startup.
docker run -p 8080:80 \
-e "NONUSE=reports" \
bgruening/galaxy-stableGalaxy's config settings [toc]
Every Galaxy configuration parameter in config/galaxy.ini can be overwritten by passing an environment variable to the docker run command during startup. The name of the environment variable has to be:
GALAXY_CONFIG+ the_original_parameter_name_in_capital_letters
For example, you can set the Galaxy session timeout to 5 minutes and set your own Galaxy brand by invoking the docker run like this:
docker run -p 8080:80 \
-e "GALAXY_CONFIG_BRAND='My own Galaxy flavour'" \
-e "GALAXY_CONFIG_SESSION_DURATION=5" \
bgruening/galaxy-stableNote, that if you would like to run any of the cleanup scripts, you will need to add the following to /export/galaxy-central/config/galaxy.yml:
database_connection = postgresql://galaxy:galaxy@localhost:5432/galaxy
file_path = /export/galaxy-central/database/files
By default the admin_users and master_api_key variables are set to:
admin_users: [email protected]
master_api_key: HSNiugRFvgT574F43jZ7N9F3
Additionally Galaxy encodes various internal values that can be part of output using secret string configurable as id_secret in the config file (use 5-65 bytes long string). This prevents 'guessing' of Galaxy's internal database sequences. Example:
id_secret: d5c910cc6e32cad08599987ab64dcfae
You should change all three configuration variables above manually in /export/galaxy-central/config/galaxy.yml.
Alternatively you can pass the security configuration when running the image but please note that it is a security problem. E.g. if a tool exposes all env's your secret API key will also be exposed.
Configuring Galaxy's behind a proxy [toc]
If your Galaxy docker instance is running behind an HTTP proxy server, and if you're accessing it with a specific path prefix (e.g. http://www.example.org/some/prefix/), you need to make Galaxy aware of it. There is an environment variable available to do so:
PROXY_PREFIX=/some/prefix
You can and should overwrite these during launching your container:
docker run -p 8080:80 \
-e "PROXY_PREFIX=/some/prefix" \
bgruening/galaxy-stableOn-demand reference data with CVMFS [toc]
By default, Galaxy instances launched with this image will have on-demand access to approximately 3TB of
reference genomes and indexes. These are the same reference data available on the main Galaxy server.
This is achieved by connecting to Galaxy's CernVM filesystem (CVMFS) at data.galaxyproject.org repository,
which is geographically distributed among numerous servers.
The CVMFS capability doesn't add to the size of the Docker image, but when running, CVMFS maintains
a cache to keep the most recently used data on the local disk.
Note: for CVMFS directories to be mounted-on-demand with autofs, you must launch Docker as --privileged
Personalize your Galaxy [toc]
The Galaxy welcome screen can be changed by providing a welcome.html page in /home/user/galaxy_storage/. All files starting with welcome will be copied during startup and served as introduction page. If you want to include images or other media, name them welcome_* and link them relative to your welcome.html (example).
Deactivating services [toc]
Non-essential services can be deactivated during startup. Set the environment variable NONUSE to a comma separated list of services. Currently, nodejs, postgres, proftp, reports, slurmd and slurmctld are supported.
docker run -d -p 8080:80 -p 9002:9002 \
-e "NONUSE=nodejs,proftp,reports,slurmd,slurmctld" \
bgruening/galaxy-stableA graphical user interface, to start and stop your services, is available on port 9002 if you run your container like above.
Restarting Galaxy [toc]
If you want to restart Galaxy without restarting the entire Galaxy container you can use docker exec (docker > 1.3).
docker exec <container name> supervisorctl restart galaxy:In addition you can start/stop every supervisord process using a web interface on port 9002. Start your container with:
docker run -p 9002:9002 bgruening/galaxy-stableAdvanced Logging [toc]
You can set the environment variable $GALAXY_LOGGING to FULL to access all logs from supervisor. For example start your container with:
docker run -d -p 8080:80 -p 8021:21 \
-e "GALAXY_LOGGING=full" \
bgruening/galaxy-stableThen, you can access the supervisord web interface on port 9002 and get access to log files. To do so, start your container with:
docker run -d -p 8080:80 -p 8021:21 -p 9002:9002 \
-e "GALAXY_LOGGING=full" \
bgruening/galaxy-stableAlternatively, you can access the container directly using the following command:
docker exec -it <container name> bashOnce connected to the container, log files are available in /home/galaxy/logs.
A volume can also be used to map this directory to one external to the container - for instance if logs need to be persisted for auditing reasons (security, debugging, performance testing, etc...).:
mkdir gx_logs
docker run -d -p 8080:80 -p 8021:21 -e "GALAXY_LOGGING=full" -v `pwd`/gx_logs:/home/galaxy/logs bgruening/galaxy-stableRunning on an external cluster (DRM) [toc]
Basic setup for the filesystem [toc]
The easiest way is to create a /export mount point on the cluster and mount the container with /export:/export.
The docker container sets up all its files on the /export directory, but this directory may not exist on the cluster filesystem. This can be solved with symbolic links on the cluster filesystem but it can also be solved within the container itself.
In this example configuration the cluster file system has a directory /cluster_storage/galaxy which is accessible for the galaxy user in the container (UID 1450) and the user starting the container.
The container should be started with the following settings configured:
docker run -d -p 8080:80 -p 8021:21 \
-v /cluster_storage/galaxy/galaxy_export:/export \ # This makes sure all galaxy files are on the cluster filesystem
-v /cluster_storage/galaxy:/cluster_storage/galaxy \ # This ensures the links within the docker container and on the cluster fs are the same
# The following settings make sure that each job is configured with the paths on the cluster fs instead of /export
-e GALAXY_CONFIG_TOOL_DEPENDENCY_DIR="/cluster_storage/galaxy/galaxy_export/tool_deps" \
-e GALAXY_CONFIG_TOOL_DEPENDENCY_CACHE_DIR="/cluster_storage/galaxy/galaxy_export/tool_deps/_cache" \
-e GALAXY_CONFIG_FILE_PATH="/cluster_storage/galaxy/galaxy_export/galaxy-central/database/files" \
-e GALAXY_CONFIG_TOOL_PATH="/cluster_storage/galaxy/galaxy_export/galaxy-central/tools" \
-e GALAXY_CONFIG_TOOL_DATA_PATH="/cluster_storage/galaxy/galaxy_export/galaxy-central/tool-data" \
-e GALAXY_CONFIG_SHED_TOOL_DATA_PATH="/cluster_storage/galaxy/galaxy_export/galaxy-central/tool-data" \
# The following settings are for directories that can be anywhere on the cluster fs.
GALAXY_CONFIG_JOB_WORKING_DIRECTORY="/cluster_storage/galaxy/galaxy_export/galaxy-central/database/job_working_directory" \ #IMPORTANT: needs to be created manually. Can also be placed elsewhere, but is originally located here
-e GALAXY_CONFIG_NEW_FILE_PATH="/cluster_storage/galaxy/tmp" \ # IMPORTANT: needs to be created manually. This needs to be writable by UID=1450 and have its flippy bit set (chmod 1777 for world-writable with flippy bit)
-e GALAXY_CONFIG_CLUSTER_FILES_DIRECTORY="/cluster_storage/galaxy/job_scripts" \ # Job scripts and stdout and stderr will be written here.
-e GALAXY_CONFIG_OUTPUTS_TO_WORKING_DIRECTORY=False \ # Writes Job scripts, stdout and stderr to job_working_directory.
-e GALAXY_CONFIG_RETRY_JOB_OUTPUT_COLLECTION=5 \ #IF your cluster fs uses nfs this may introduce latency. You can set galaxy to retry if a job output is not yet created.
# Conda settings. IMPORTANT!
-e GALAXY_CONFIG_CONDA_PREFIX="/cluster_storage/galaxy/_conda" \ # Can be anywhere EXCEPT cluster_storage/galaxy/galaxy_export!
# Conda uses $PWD to determine where the virtual environment is. If placed inside the export directory conda will determine $PWD to be a subirectory of the /export folder which does not exist on the cluster!
-e GALAXY_CONFIG_CONDA_AUTO_INIT=True # When the necessary environment can not be found a new one will automatically be createdSetting up a Python virtual environment on the cluster [toc]
The Python environment in the container is not accessible from the cluster. So it needs to be created beforehand.
In this example configuration the Python virtual environment is created on /cluster_storage/galaxy/galaxy_venv and the export folder on /cluster_storage/galaxy/galaxy_export. To create the virtual environment:
- Create the virtual environment
virtualenv /cluster_storage/galaxy/galaxy_venv - Activate the virtual environment
source /cluster_storage/galaxy/galaxy_venv/bin/activate - Install the galaxy requirements
pip install --index-url https://wheels.galaxyproject.org/simple --only-binary all -r /cluster_storage/galaxy/galaxy-central//lib/galaxy/dependencies/pinned-requirements.txt
- Make sure to upgrade the environment with the new requirements when a new version of galaxy is released.
To make the Python environment usable on the cluster, create your custom job_conf.xml file and put it in /cluster_storage/galaxy/galaxy_export/galaxy-central/config.
In the destination section the following code should be added:
<destinations default="cluster">
<destination id="cluster" runner="your_cluster_runner">
<env file="/cluster_storage/galaxy/galaxy_venv/bin/activate"/>
<env id="GALAXY_ROOT_DIR">/cluster_storage/galaxy/galaxy_export/galaxy-central</env>
<env id="GALAXY_LIB">/cluster_storage/galaxy/galaxy_export/galaxy-central/lib</env>
<env id="PYTHONPATH">/cluster_storage/galaxy/galaxy_export/galaxy-central/lib</env>
<param id="embed_metadata_in_job">True</param>
</destination>In this way, Python tools on the cluster are able to use the Galaxy libraries.
More information can be found here and here.
Using an external Slurm cluster [toc]
It is often convenient to configure Galaxy to use a high-performance cluster for running jobs. To do so, two files are required:
- munge.key
- slurm.conf
These files from the cluster must be copied to the /export mount point (i.e., /cluster_storage/galaxy/galaxy_export/ on the host if using below command) accessible to Galaxy before starting the container. This must be done regardless of which Slurm daemons are running within Docker. At start, symbolic links will be created to these files to /etc within the container, allowing the various Slurm functions to communicate properly with your cluster. In such cases, there's no reason to run slurmctld, the Slurm controller daemon, from within Docker, so specify -e "NONUSE=slurmctld". Unless you would like to also use Slurm (rather than the local job runner) to run jobs within the Docker container, then alternatively specify -e "NONUSE=slurmctld,slurmd".
Importantly, Slurm relies on a shared filesystem between the Docker container and the execution nodes. To allow things to function correctly, checkout the basic filesystem setup above.
A brief note is in order regarding the version of Slurm installed. This Docker image uses Ubuntu 14.04 as its base image. The version of Slurm in the Ubuntu 14.04 repository is 2.6.5 and that is what is installed in this image. If your cluster is using an incompatible version of Slurm then you will likely need to modify this Docker image.
The following is an example for how to specify a destination in job_conf.xml that uses a custom partition ("work", rather than "debug") and 4 cores rather than 1:
<destination id="slurm4threads" runner="slurm">
<param id="embed_metadata_in_job">False</param>
<param id="nativeSpecification">-p work -n 4</param>
</destination>
The usage of -n can be confusing. Note that it will specify the number of cores, not the number of tasks (i.e., it's not equivalent to srun -n 4).
Using an external Grid Engine cluster [toc]
Set up the filesystem on the cluster as mentioned above. To use Grid Engine (Sun Grid Engine, Open Grid Scheduler), one configuration file and an environment variable are required:
- create an
act_qmasterfile in the /export folder.
- In act_qmaster is something like this.
YOUR_GRIDENGINE_MASTER_HOST
- this file will automatically be installed in the container's
/var/lib/gridenginefolder.
- set the environment variable
SGE_ROOT
- By default
-e SGE_ROOT=/var/lib/gridengine
- Make sure that YOUR_GRIDENGINE_MASTER_HOST can be pinged from the docker container. If this is not the case you can put the qmaster's hostname and ip in the containers
/etc/hostsYour Grid Engine needs to accept job submissions from inside the container. If your container is already on a host that can submit jobs, set the hostname of the container to be exactly the same as the host. (The hostname can be changed by using the --hostname flag when starting the container).
Alternatively, you can add the container's hostname (default=galaxy-docker) to the /etc/hosts file on the gridengine head node. And setting the container's hostname as a submit host.
Tips for Running Jobs Outside the Container [toc]
In its default state Galaxy assumes both the Galaxy source code and various temporary files are available on shared file systems across the cluster. When using Condor or SLURM (as described above) to run jobs outside of the Docker container one can take steps to mitigate these assumptions.
The embed_metadata_in_job option on job destinations in job_conf.xml
forces Galaxy collect metadata inside the container instead of on the
cluster:
<param id="embed_metadata_in_job">False</param>
This has performance implications and may not scale as well as performing these calculations on the remote cluster - but this should not be a problem for most Galaxy instances.
Enable Galaxy to use BioContainers (Docker) [toc]
This is a very cool feature where Galaxy automatically detects that your tool has an associated docker image, pulls it and runs it for you. These images (when available) have been generated using mulled. To test, install the IUC bedtools from the toolshed. When you try to execute ClusterBed for example. You may get a missing dependancy error for bedtools. But bedtools has an associated docker image on quay.io. Now configure Galaxy as follows:
- Add this environment variable to
docker run:-e GALAXY_CONFIG_ENABLE_BETA_MULLED_CONTAINERS=True - In
job_conf.xmlconfigure a Docker enabled destination as follows:
<destination id="docker_local" runner="local">
<param id="docker_enabled">true</param>
<param id="docker_volumes">$galaxy_root:ro,$galaxy_root/database/tmp:rw,$tool_directory:ro,$job_directory:ro,$working_directory:rw,$default_file_path:rw</param>
<param id="docker_sudo">false</param>
</destination>When you execute the tool again, Galaxy will pull the image from Biocontainers (quay.io/biocontainers), run the tool inside of this container to produce the desired output.
Magic Environment variables [toc]
| Name | Description |
|---|---|
ENABLE_TTS_INSTALL |
Enables the Test Tool Shed during container startup. This change is not persistent. (ENABLE_TTS_INSTALL=True) |
GALAXY_LOGGING |
Enables for verbose logging at Docker stdout. (GALAXY_LOGGING=full) |
BARE |
Disables all default Galaxy tools. (BARE=True) |
NONUSE |
Disable services during container startup. (NONUSE=nodejs,proftp,reports,slurmd,slurmctld) |
UWSGI_PROCESSES |
Set the number of uwsgi processes (`UWSGI_PROCESSES=2) |
UWSGI_THREADS |
Set the number of uwsgi threads (UWSGI_THREADS=4) |
GALAXY_DOCKER_ENABLED |
Enable Galaxy to use Docker containers if annotated in tools (GALAXY_DOCKER_ENABLED=False) |
GALAXY_DOCKER_VOLUMES |
Specify volumes that should be mounted into tool containers (GALAXY_DOCKER_VOLUMES="") |
GALAXY_HANDLER_NUMPROCS |
Set the number of Galaxy handler (GALAXY_HANDLER_NUMPROCS=2) |
LOAD_GALAXY_CONDITIONAL_DEPENDENCIES |
Installing optional dependencies into the Galaxy virtual environment |
LOAD_PYTHON_DEV_DEPENDENCIES |
Installation of Galaxy's dev dependencies. Needs LOAD_GALAXY_CONDITIONAL_DEPENDENCIES as well |
HTTPS Support [toc]
It's possible to automatically configure your container with HTTPS, either with certificates of your own or by automatically requesting an HTTPS certificate from Letsencrypt with the following environment variables:
| Name | Description |
|---|---|
USE_HTTPS |
Set USE_HTTPS=True to set up HTTPS via self-signed certificates. If you have your own certificates, copy them to /export/{server.key,server.crt}. |
USE_HTTPS_LETSENCRYPT |
Set USE_HTTPS_LETSENCRYPT=True to automatically set up HTTPS using Letsencrypt as a certificate authority. (Requires you to also set GALAXY_CONFIG_GALAXY_INFRASTRUCTURE_URL) Note: only set one of USE_HTTPS and USE_HTTPS_LETSENCRYPT to true. |
GALAXY_CONFIG_GALAXY_INFRASTRUCTURE_URL |
Set GALAXY_CONFIG_GALAXY_INFRASTRUCTURE_URL=<your_domain> so that Letsencrypt can test your that you own the domain you claim to own in order to issue you your HTTPS cert. |
Lite Mode [toc]
The lite mode will only start postgresql and a single Galaxy process, without nginx, uwsgi or any other special feature from the normal mode. In particular there is no support for the export folder or any Magic Environment variables.
docker run -i -t -p 8080:8080 bgruening/galaxy-stable startup_liteThis will also use the standard job_conf.xml.sample_basic shipped by Galaxy. If you want to use the regular one from the normal mode you can pass -j to the startup_lite script.
Extending the Docker Image [toc]
If the desired tools are already included in the Tool Shed, building your own personalised Galaxy docker Image (Galaxy flavour) can be done using the following steps:
- Create a file named
Dockerfile - Include
FROM bgruening/galaxy-stableat the top of the file. This means that you use the Galaxy Docker Image as base Image and build your own extensions on top of it. - Supply the list of desired tools in a file (
my_tool_list.ymlbelow). See this page for the file format requirements. - Execute
docker build -t my-docker-test .4a. (if behind proxy). Add the ENV http_proxy and https_proxy variables as IPs (to avoid nameserver resolution problems) as in the example below. - Run your container with
docker run -p 8080:80 my-docker-test - Open your web browser on
http://localhost:8080
For a working example, have a look at these Dockerfiles.
# Galaxy - deepTools
#
# VERSION 0.2
FROM bgruening/galaxy-stable
MAINTAINER Björn A. Grüning, [email protected]
ENV GALAXY_CONFIG_BRAND deepTools
# The following two lines are optional and can be given during runtime
# with the -e http_proxy='http://yourproxyIP:8080' parameter
ENV http_proxy 'http://yourproxyIP:8080'
ENV https_proxy 'http://yourproxyIP:8080'
WORKDIR /galaxy-central
RUN add-tool-shed --url 'http://testtoolshed.g2.bx.psu.edu/' --name 'Test Tool Shed'
# Install Visualisation
RUN install-biojs msa
# Adding the tool definitions to the container
ADD my_tool_list.yml $GALAXY_ROOT/my_tool_list.yml
# Install deepTools
RUN install-tools $GALAXY_ROOT/my_tool_list.yml
# Mark folders as imported from the host.
VOLUME ["/export/", "/data/", "/var/lib/docker"]
# Expose port 80 (webserver), 21 (FTP server), 8800 (Proxy)
EXPOSE :80
EXPOSE :21
EXPOSE :8800
# Autostart script that is invoked during container start
CMD ["/usr/bin/startup"]
or the RNA-workbench. The RNA-workbench has advanced examples about:
-
populating Galaxy data libraries
setup-data-libraries -i $GALAXY_ROOT/library_data.yaml -g http://localhost:8080 -u $GALAXY_DEFAULT_ADMIN_USER -p $GALAXY_DEFAULT_ADMIN_PASSWORD
The actual data is references in a YAML file similar this one.
-
installing workflows
workflow-install --workflow_path $GALAXY_HOME/workflows/ -g http://localhost:8080 -u $GALAXY_DEFAULT_ADMIN_USER -p $GALAXY_DEFAULT_ADMIN_PASSWORD
Where all Galaxy workflows needs to be in one directory, here the $GALAXY_HOME/workflows/.
-
running Galaxy data-managers to create indices or download data
run-data-managers -u $GALAXY_DEFAULT_ADMIN_USER -p $GALAXY_DEFAULT_ADMIN_PASSWORD -g http://localhost:8080 --config data_manager_rna_seq.yaml
The data-managers can be configured and specified in a YAML file similar to this one.
If you host your flavor on GitHub consider to test our build with Travis-CI. This project will help you: https://github.com/bgruening/galaxy-flavor-testing
List of Galaxy flavours [toc]
- Aurora Galaxy
- SNP analysis Workflows on Docker (sniplay)
- NCBI-Blast
- ChemicalToolBox
- ballaxy
- NGS-deepTools
- Galaxy ChIP-exo
- Galaxy Proteomics
- Imaging
- Constructive Solid Geometry
- Galaxy for metagenomics
- Galaxy with the Language Application Grid tools
- RNAcommender
- OpenMoleculeGenerator
- Workflow4Metabolomics
- HiC-Explorer
- SNVPhyl
- GraphClust
- RNA workbench
- Cancer Genomics Toolkit
- Clustered Heatmaps for Interactive Exploration of Molecular Profiling Data
Integrating non-Tool Shed tools into the container [toc]
We recommend to use the Main Galaxy Tool Shed for all your tools and workflows that you would like to share. In rare situations where you cannot share your tools but still want to include them into your Galaxy Docker instance, please follow the next steps.
-
Get your tools into the container.
Mount your tool directory into the container with a separate
-v /home/user/my_galaxy_tools/:/local_tools. -
Create a
tool_conf.xmlfile for your tools.This should look similar to the main
tool_conf.xmlfile, but references your tools from the new directory. In other words a tool entry should look like this<tool file="/local_tools/application_foo/foo.xml" />. Yourtool_conf.xmlshould be available from inside of the container. We assume you have it stored under/local_tools/my_tools.xml. -
Add the new tool config file to the Galaxy configuration.
To make Galaxy aware of your new tool configuration file you need to add the path to
tool_config_file, which is by default#tool_config_file = config/tool_conf.xml,config/shed_tool_conf.xml. You can do this during container start by setting the environment variable-e GALAXY_CONFIG_TOOL_CONFIG_FILE=config/tool_conf.xml.sample,config/shed_tool_conf.xml.sample,/local_tools/my_tools.xml.
Users & Passwords [toc]
The Galaxy Admin User has the username [email protected] and the password password.
The PostgreSQL username is galaxy, the password is galaxy and the database name is galaxy (I know I was really creative ;)).
If you want to create new users, please make sure to use the /export/ volume. Otherwise your user will be removed after your docker session is finished.
The proftpd server is configured to use the main galaxy PostgreSQL user to access the database and select the username and password. If you want to run the
docker container in production, please do not forget to change the user credentials in /etc/proftpd/proftpd.conf too.
The Galaxy Report Webapp is htpasswd protected with username and password set to admin.
Development [toc]
This repository uses a git submodule to include Ansible roles maintained by the Galaxy project.
You can clone this repository and the Ansible submodule with:
git clone --recursive https://github.com/bgruening/docker-galaxy-stable.gitUpdating already existing submodules is possible with:
git submodule update --init --recursiveIf you simply want to change the Galaxy repository and/or the Galaxy branch, from which the container is build you can do this with Docker --build-arg during the docker build step. For example you can use these parameters during container build:
--build-arg GALAXY_RELEASE=install_workflow_and_tools
--build-arg GALAXY_REPO=https://github.com/manabuishii/galaxy
Requirements [toc]
History [toc]
- 0.1: Initial release!
- with Apache2, PostgreSQL and Tool Shed integration
- 0.2: complete new Galaxy stack.
- with nginx, uwsgi, proftpd, docker, supervisord and SLURM
- 0.3: Add Interactive Environments
- IPython in docker in Galaxy in docker
- advanged logging
- 0.4:
- base the image on toolshed/requirements with all required Galaxy dependencies
- use Ansible roles to build large parts of the image
- export the supervisord web interface on port 9002
- enable Galaxy reports webapp
- 15.07:
install-biojscan install BioJS visualisations into Galaxyadd-tool-shedcan be used to activate third party Tool Sheds in child Dockerfiles- many documentation improvements
- RStudio is now part of Galaxy and this Image
- configurable postgres UID/GID by @chambm
- smarter starting of postgres during Tool installations by @shiltemann
- 15.10:
- new Galaxy 15.10 release
- fix #94
- 16.01:
- enable Travis testing for all builds and PR
- offer new yaml based tool installations
- enable dynamic UWSGI processes and threads with
-e UWSGI_PROCESSES=2and-e UWSGI_THREADS=4 - enable dynamic Galaxy handlers
-e GALAXY_HANDLER_NUMPROCS=2 - Addition of a new
litemode contributed by @kellrott - first release with Jupyter integration
- 16.04:
- include a Galaxy-bare mode, enable with
-e BARE=True - first release with HTCondor installed and pre-configured
- include a Galaxy-bare mode, enable with
- 16.07:
- documentation and tests updates for SLURM integration by @mvdbeek
- first version with initial Docker compose support (proftpd ✔️)
- SFTP support by @zfrenchee
- 16.10:
- HTTPS support by @zfrenchee and @mvdbeek
- 17.01:
- enable Conda dependency resolution by default
- new Galaxy version
- more compose work (slurm, postgresql)
- 17.05:
- add PROXY_PREFIX variable to enable automatic configuration of Galaxy running under some prefix (@abretaud)
- enable quota by default (just the funtionality, not any specific value)
- HT-Condor is now supported in compose with semi-autoscaling and BioContainers
- Galaxy Docker Compose is completely under Travis testing and available with SLURM and HT-Condor
- using Docker
build-args for GALAXY_RELEASE and GALAXY_REPO
- 17.09:
- much improved documentation about using Galaxy Docker and an external cluster (@rhpvorderman)
- CVMFS support - mounting in 4TB of pre-build reference data (@chambm)
- Singularity support and tests (compose only)
- more work on K8s support and testing (@jmchilton)
- using .env files to configure the compose setup for SLURM, Condor, K8s, SLURM-Singularity, Condor-Docker
- 18.01:
- tracking the Galaxy release_18.01 branch
- uwsgi work to adopt to changes for 18.01
- remove nodejs-legacy & npm from Dockerfile and install latest version from ansible-extras
- initial galaxy.ini → galaxy.yml integration
- grafana and influxdb container (compose)
- Galaxy telegraf integration to push to influxdb (compose)
- added some documentation (compose)
- 18.05:
- Nothing very special, but a awesome Galaxy release as usual
- 18.09:
- new and more powerful orchestration build script (build-orchestration-images.sh) by @pcm32
- a lot of bug-fixes to the compose setup by @abretaud
- 19.01:
- This is featuring the latest and greatest from the Galaxy community
- Please note that this release will be the last release which is based on
ubuntu:14.04and PostgreSQL 9.3. We will migrate toubuntu:18.04and a newer PostgreSQL version in19.05. Furthermore, we will not support old Galaxy tool dependencies.
- 19.05:
- The image is now based on
ubuntu:18.04(instead of ubuntu:14.04 previously) and PostgreSQL 11.5 (9.3 previously). See migration documention to migrate the postgresql database from 9.3 to 11.5. - We not longer support old Galaxy tool dependencies.
- The image is now based on
- 20.05:
- Featuring Galaxy 20.05
- Completely reworked compose setup
- The default admin password and apikey (
GALAXY_DEFAULT_ADMIN_PASSWORDandGALAXY_DEFAULT_ADMIN_KEY) have changed: the password is nowpassword(instead ofadmin) and the apikeyfakekey(instead ofadmin).
- 20.09:
- Featuring Galaxy 20.09
Support & Bug Reports [toc]
You can file an github issue or ask us on the Galaxy development list.
If you like this service please fill out this survey: https://www.surveymonkey.de/r/denbi-service?sc=rbc&tool=galaxy-docker