karchinlab / open-cravat Goto Github PK
View Code? Open in Web Editor NEWA modular annotation tool for genomic variants
License: MIT License
A modular annotation tool for genomic variants
License: MIT License
Hello,
After waiting ~3 hours and being exciting to see the results :( ; I got an error when I'm trying to download the results with excel/text format and when I investigate the log file; it showed me this:
2019/07/23 00:01:09 cravat [Errno 13] Permission denied: '/Applications/OpenCRAVAT.app/Contents/Resources/dummy.log'
Traceback (most recent call last):
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/cravat_class.py", line 673, in run_reporter
await reporter.run()
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/cravat_report.py", line 146, in run
await self.make_col_info(level)
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/cravat_report.py", line 348, in make_col_info
annot = annot_cls([mi.script_path, 'dummy'], {})
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_annotator.py", line 76, in init
self._log_exception(e)
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_annotator.py", line 80, in _log_exception
raise e
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_annotator.py", line 58, in init
self._setup_logger()
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_annotator.py", line 449, in _setup_logger
self._log_exception(e)
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_annotator.py", line 80, in _log_exception
raise e
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_annotator.py", line 438, in _setup_logger
log_handler = logging.FileHandler(self.log_path, 'a')
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/logging/init.py", line 1092, in init
StreamHandler.init(self, self._open())
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/logging/init.py", line 1121, in _open
return open(self.baseFilename, self.mode, encoding=self.encoding)
PermissionError: [Errno 13] Permission denied: '/Applications/OpenCRAVAT.app/Contents/Resources/dummy.log'
2019/07/23 00:01:09 cravat finished with an exception: Tue Jul 23 00:01:09 2019
2019/07/23 00:01:09 cravat runtime: 0.181s
2019/07/23 00:01:14 cravat started: Tue Jul 23 00:01:14 2019
2019/07/23 00:01:14 cravat input assembly: hg38
2019/07/23 00:01:15 cravat.excelreporter started: Tue Jul 23 00:01:15 2019
2019/07/23 00:01:15 cravat [Errno 13] Permission denied: '/Applications/OpenCRAVAT.app/Contents/Resources/dummy.log'
Traceback (most recent call last):
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/cravat_class.py", line 673, in run_reporter
await reporter.run()
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/cravat_report.py", line 146, in run
await self.make_col_info(level)
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/cravat_report.py", line 348, in make_col_info
annot = annot_cls([mi.script_path, 'dummy'], {})
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_annotator.py", line 76, in init
self._log_exception(e)
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_annotator.py", line 80, in _log_exception
raise e
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_annotator.py", line 58, in init
self._setup_logger()
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_annotator.py", line 449, in _setup_logger
self._log_exception(e)
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_annotator.py", line 80, in _log_exception
raise e
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_annotator.py", line 438, in _setup_logger
log_handler = logging.FileHandler(self.log_path, 'a')
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/logging/init.py", line 1092, in init
StreamHandler.init(self, self._open())
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/logging/init.py", line 1121, in _open
return open(self.baseFilename, self.mode, encoding=self.encoding)
PermissionError: [Errno 13] Permission denied: '/Applications/OpenCRAVAT.app/Contents/Resources/dummy.log'
test
We are trying to use open-cravat from remote computer.
Host computer OS where open-cravat is installed: Windows 10 Enterprise
Host computer name where open-cravat is installed: cravat
Host computer domain: example.com
For that we installed multiuser support via WinPython Command Prompt as admin:
pip install open-cravat-multiuser
After that we changed open-cravat config:
Config file location: C:\open-cravat\conf\cravat.yml
converter: converter
genemapper: hg38
aggregator: aggregator
reporter: excelreporter
gui_host: www.cravat.example.com
gui_port: 80
After that we opened firewall port for TCP 80 from cravat host.
Now run open-cravat from WinPython Command Prompt as admin:
wcravat --multiuser --donotopenbrowser
No matter what gui_host or gui_port is defined output is same:
C:\Program Files (x86)\open-cravat\scripts>wcravat --multiuser --donotopenbrowser
OpenCRAVAT is served at localhost:8060
(To quit: Press Ctrl-C or Ctrl-Break if run on a Terminal or Windows, or click "Cancel" and then "Quit" if run through OpenCRAVAT app on Mac OS)
open-cravat is running only in localhost:8060 and is inaccessible from remote computers as http://cravat.example.com
What im i missing?
conda create --name cravat
conda activate cravat
conda install pip
pip install open-cravat
oc module install --yes clinvar vcfreporter vcf-converter hg38
wget https://raw.githubusercontent.com/vcflib/vcflib/master/samples/sample.vcf
oc run sample.vcf --repeat converter -t vcf -l hg38
Running reporter...
VCF Reporter (vcfreporter) Traceback (most recent call last):
File "/opt/anaconda3/envs/cravat/lib/python3.7/site-packages/cravat/cravat_class.py", line 875, in run_reporter
await reporter.run()
File "/opt/anaconda3/envs/cravat/lib/python3.7/site-packages/cravat/cravat_report.py", line 354, in run
await self.run_level(level)
File "/opt/anaconda3/envs/cravat/lib/python3.7/site-packages/cravat/cravat_report.py", line 219, in run_level
gene_summary_data = await o.get_gene_summary_data(self.cf)
File "/opt/anaconda3/envs/cravat/lib/python3.7/site-packages/cravat/base_mapper.py", line 288, in get_gene_summary_data
rows = await cf.get_variant_data_for_cols(cols)
File "/opt/anaconda3/envs/cravat/lib/python3.7/site-packages/cravat/cravat_filter.py", line 676, in get_variant_data_for_cols
await self.cursor.execute(q)
File "/opt/anaconda3/envs/cravat/lib/python3.7/site-packages/aiosqlite3/cursor.py", line 130, in execute
res = yield from self._execute(self._cursor.execute, sql, parameters)
File "/opt/anaconda3/envs/cravat/lib/python3.7/site-packages/aiosqlite3/cursor.py", line 56, in _execute
res = yield from self._conn.async_execute(func, *args, **kwargs)
File "/opt/anaconda3/envs/cravat/lib/python3.7/site-packages/aiosqlite3/connection.py", line 137, in async_execute
return (yield from self._execute(func, *args, **kwargs))
File "/opt/anaconda3/envs/cravat/lib/python3.7/site-packages/aiosqlite3/connection.py", line 128, in _execute
func
File "/opt/anaconda3/envs/cravat/lib/python3.7/concurrent/futures/thread.py", line 57, in run
result = self.fn(*self.args, **self.kwargs)
sqlite3.OperationalError: no such column: tagsampler__numsample
Hm - this is tricky:
To hide the yaml warnings I had redirected stderr in a wrapper script. After install of 1.8.0, ocwrapper module update listed four things to update. Then nothing. For a really long time - until I clued in. Not sure I agree with the use of both stdout and stderr in the same breath?
Then a slew of fun of our own making - a nasty permissions thing. Redo the install/update.Then a trial -expect nothing - run of oc module update:
oc module update
Newer versions of (cravat-converter, oldcravat-converter, vcf-converter) are available, but would break dependencies. You may use --strategy=force to force installation.
No module updates are needed
I think we're good, but a little shakey. (I don't use oc, I'll pester she-who-does to try it asap)
Originally posted by @iceback in #26 (comment)
Issue: Store and jobs disappear on webpage with recent system moduleupdates.
Log shows:
_File "C:\Program Files (x86)\open-cravat\python-3.7.2.amd64\lib\site-packages\cravat\cravat_web.py", line 283, in middleware
response = await handler(request)
File "C:\Program Files (x86)\open-cravat\python-3.7.2.amd64\lib\site-packages\cravat\websubmit\websubmit.py", line 598, in get_report_types
valid_types = get_valid_report_types()
File "C:\Program Files (x86)\open-cravat\python-3.7.2.amd64\lib\site-packages\cravat\websubmit\websubmit.py", line 589, in get_valid_report_types
reporter_infos = au.get_local_module_infos(types=['reporter'])
File "C:\Program Files (x86)\open-cravat\python-3.7.2.amd64\lib\site-packages\cravat\admin_util.py", line 357, in get_local_module_infos
all_infos = list(mic.local.values())
File "C:\Program Files (x86)\open-cravat\python-3.7.2.amd64\lib_collections_abc.py", line 762, in iter
yield self.mapping[key]
File "C:\Program Files (x86)\open-cravat\python-3.7.2.amd64\lib\site-packages\cravat\admin_util.py", line 198, in getitem
self.store[key] = LocalModuleInfo(self.store[key])
File "C:\Program Files (x86)\open-cravat\python-3.7.2.amd64\lib\site-packages\cravat\admin_util.py", line 91, in init
self.readme = f.read()
File "C:\Program Files (x86)\open-cravat\python-3.7.2.amd64\lib\encodings\cp1252.py", line 23, in decode
return codecs.charmap_decode(input,self.errors,decoding_table)[0]
UnicodeDecodeError: 'charmap' codec can't decode byte 0x81 in position 1806: character maps to
This is caused by lines 90 & 91 in admin_util.py, encoding should be set explicitely to UTF-8 as follows:
if self.readme_exists:
with open(self.readme_path, encoding='utf-8') as f:
self.readme = f.read()
If I'm interested in the .sqlite, but don't want the .tsv nor the .xlsx is there a way to avoid making both of them?
test
Currently the wiki shows https://pypi.org/project/CRAVAT/
but it should be
I just did a database update, and find that all strings that formerly contained spaces, now have those spaces replaced by underscores.
affected fields include at least
clinvar__disease_names : example 'Argininosuccinate lyase deficiency' -> 'Argininosuccinate_lyase_deficiency'
clinvar__disease_refs : where the database labels 'SNOMED CT' -> 'SNOMED_CT'. The same is true elsewhere for the databases names Human_Phenotype_Ontology
clinvar__rev_stat
clinvar__sig is also affected, where numerous strings have changed such as
"drug response" to "drug_response"
"Pathogenic/Likely pathogenic" to "Pathogenic/Likely_pathogenic"
"risk factor" to "risk_factor"
"Likely pathogenic" to "Likely_pathogenic"
etc
I could cope with either representation, but worry that this may be a bug, and might eventually be flipped back. If it's arbitrary to you, I think the spaces are preferable.
In addition to reporting the p-value, can you also please output the FDR? Thanks.
Hi open-cravat team,
I'm facing an error while open cravat-web based, this is the error after I upload my vcf file:
2019/07/22 16:22:41 cravat started: Mon Jul 22 16:22:41 2019
2019/07/22 16:22:41 cravat input assembly: hg38
2019/07/22 16:22:41 cravat.converter started: Mon Jul 22 16:22:41 2019
2019/07/22 16:22:41 cravat.converter input files: /Users/Shared/open-cravat/jobs/default/190722-162237/CosmicCodingMuts.vcf
2019/07/22 16:22:55 cravat.converter input format: vcf
2019/07/22 16:24:36 cravat.converter error lines: 0
2019/07/22 16:24:36 cravat.converter finished: Mon Jul 22 16:24:36 2019
2019/07/22 16:24:36 cravat.converter num input lines: 4668387
2019/07/22 16:24:36 cravat.converter runtime: 100.444
2019/07/22 16:24:36 cravat.mapper input file: /Users/Shared/open-cravat/jobs/default/190722-162237/CosmicCodingMuts.vcf.crv
2019/07/22 16:24:44 cravat.mapper mapper database: /Users/Shared/open-cravat/modules/mappers/hg38/data/hg38.sqlite
2019/07/22 16:24:44 cravat.mapper started: Mon Jul 22 16:24:44 2019
2019/07/22 16:25:50 cravat.mapper Traceback (most recent call last):
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_mapper.py", line 168, in run
crx_data, alt_transcripts = self.map(crv_data)
File "/Users/Shared/open-cravat/modules/mappers/hg38/hg38.py", line 80, in map
all_hits += self._get_coding_hits(crv_data)
File "/Users/Shared/open-cravat/modules/mappers/hg38/hg38.py", line 218, in _get_coding_hits
self._fill_coding_so(hit)
File "/Users/Shared/open-cravat/modules/mappers/hg38/hg38.py", line 308, in _fill_coding_so
self._fill_snv_pchange(hit)
File "/Users/Shared/open-cravat/modules/mappers/hg38/hg38.py", line 333, in _fill_snv_pchange
hit.aalt = cravat.translate_codon(hit.full_alt)
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/util.py", line 111, in translate_codon
return codon_table[bases]
KeyError: 'CNC'
2019/07/22 16:25:50 cravat An unexpected exception occurred.
Traceback (most recent call last):
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/cravat_class.py", line 313, in main
self.run_genemapper()
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/cravat_class.py", line 569, in run_genemapper
genemapper.run()
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_mapper.py", line 173, in run
self._log_runtime_error(ln, line, e)
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_mapper.py", line 252, in _log_runtime_error
raise e
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/base_mapper.py", line 168, in run
crx_data, alt_transcripts = self.map(crv_data)
File "/Users/Shared/open-cravat/modules/mappers/hg38/hg38.py", line 80, in map
all_hits += self._get_coding_hits(crv_data)
File "/Users/Shared/open-cravat/modules/mappers/hg38/hg38.py", line 218, in _get_coding_hits
self._fill_coding_so(hit)
File "/Users/Shared/open-cravat/modules/mappers/hg38/hg38.py", line 308, in _fill_coding_so
self._fill_snv_pchange(hit)
File "/Users/Shared/open-cravat/modules/mappers/hg38/hg38.py", line 333, in _fill_snv_pchange
hit.aalt = cravat.translate_codon(hit.full_alt)
File "/Applications/OpenCRAVAT.app/Contents/Resources/lib/python3.7/site-packages/cravat/util.py", line 111, in translate_codon
return codon_table[bases]
KeyError: 'CNC'
2019/07/22 16:25:50 cravat finished with an exception: Mon Jul 22 16:25:50 2019
2019/07/22 16:25:50 cravat runtime: 188.819s
2019/07/22 16:26:32 cravat started: Mon Jul 22 16:26:32 2019
2019/07/22 16:26:32 cravat input assembly: hg38
2019/07/22 16:26:32 cravat finished: Mon Jul 22 16:26:32 2019
2019/07/22 16:26:32 cravat runtime: 0.032s
It should be relatively straightforward to write the tsv to a tsv.gz via a pipe, and often there is a performance gain by reducing the amount of disk IO.
Is this feature compelling or undesirable?
Hi!
I am trying to annotate a population vcf file (75 samples) but I am getting an error. You can access the vcf file and the log in the link bellow:
https://drive.google.com/drive/folders/1jjwGNmujaNm8Wx9iaoeUIlTk7UsGQTM6?usp=sharing
Thanks for your support!
Not sure if this is the right place to ask this question. But how does open cravat deal with variants that are not in any of the listed annotation databases. Does it still determine the impact on the protein as e.g. VEP does?
Not sure if this is a user error or something else.
After running trough mutect/freebayes and hc and merging it resulted in a 277K line vcf. This is tried to run through OC but based on this error I cannot guess the error nor get an line triggering it. Running a 5K head of the vcf works fine (4.2K header lines).
What I ran:
#log for ok 5000 lines vcf
( module load open-cravat/1.8.0-foss-2018a-Python-3.6.4; mkdir -p /path/to/oc_test/; oc run TEST.vcf -d /path/to/oc_test/ --mp 4 -l hg38 -t vcf )
Genome assembly: hg38
Running converter...
Converter (converter) finished in 1.250s
Running gene mapper... finished in 3.620s
Running annotators...
hgvs: started at Tue Sep 8 10:54:44 2020
clingen: started at Tue Sep 8 10:54:44 2020
clinvar: started at Tue Sep 8 10:54:44 2020
dbsnp: started at Tue Sep 8 10:54:44 2020
clingen: finished at Tue Sep 8 10:54:44 2020
clingen: runtime 0.101s
interpro: started at Tue Sep 8 10:54:44 2020
clinvar: finished at Tue Sep 8 10:54:45 2020
clinvar: runtime 1.420s
gnomad: started at Tue Sep 8 10:54:45 2020
interpro: finished at Tue Sep 8 10:54:46 2020
interpro: runtime 2.137s
mutation_assessor: started at Tue Sep 8 10:54:46 2020
dbsnp: finished at Tue Sep 8 10:54:47 2020
dbsnp: runtime 2.770s
cadd_exome: started at Tue Sep 8 10:54:47 2020
mutation_assessor: finished at Tue Sep 8 10:54:48 2020
mutation_assessor: runtime 2.116s
cosmic: started at Tue Sep 8 10:54:48 2020
cadd_exome: finished at Tue Sep 8 10:54:48 2020
cadd_exome: runtime 1.622s
gnomad: finished at Tue Sep 8 10:54:49 2020
gnomad: runtime 3.908s
hgvs: finished at Tue Sep 8 10:54:50 2020
hgvs: runtime 6.011s
cosmic: finished at Tue Sep 8 10:54:50 2020
cosmic: runtime 1.623s
annotator(s) finished in 7.567s
Running aggregator...
Variants finished in 3.336s
Genes finished in 0.147s
Samples finished in 0.170s
Tags finished in 0.267s
Running postaggregators...
Variant Metadata (varmeta) finished in 0.021s
Tag Sampler (tagsampler) finished in 0.576s
VCF Info (vcfinfo) finished in 0.655s
Running reporter...
VCF Reporter (vcfreporter) interpro: getting gene summary data
interpro: finished getting gene summary data in 0.001s
finished in 5.398s
Finished normally. Runtime: 23.583s
Error:
( module load open-cravat/1.8.0-foss-2018a-Python-3.6.4; mkdir -p /path/to/oc/; oc run /path/to/data.vcf -d /path/to/oc/ --mp 4 -l hg38 -t vcf )
***snip***
Running gene mapper... finished in 27.621s
Running annotators...
hgvs: started at Tue Sep 8 11:30:21 2020
clingen: started at Tue Sep 8 11:30:21 2020
gnomad: started at Tue Sep 8 11:30:21 2020
clinvar: started at Tue Sep 8 11:30:21 2020
clingen: finished at Tue Sep 8 11:30:21 2020
clingen: runtime 0.204s
interpro: started at Tue Sep 8 11:30:21 2020
clinvar: finished at Tue Sep 8 11:35:10 2020
clinvar: runtime 289.050s
dbsnp: started at Tue Sep 8 11:35:10 2020
interpro: finished at Tue Sep 8 11:37:27 2020
interpro: runtime 426.401s
mutation_assessor: started at Tue Sep 8 11:37:27 2020
umcg-mterpstra@pg-interactive:molgenis-c5-TumorNormal gnomad: finished at Tue Sep 8 11:43:18 2020
gnomad: runtime 776.939s
cadd_exome: started at Tue Sep 8 11:43:18 2020
mutation_assessor: finished at Tue Sep 8 11:45:08 2020
mutation_assessor: runtime 460.258s
cosmic: started at Tue Sep 8 11:45:08 2020
cadd_exome: finished at Tue Sep 8 11:50:07 2020
cadd_exome: runtime 409.182s
cosmic: finished at Tue Sep 8 11:51:31 2020
cosmic: runtime 383.582s
hgvs: finished at Tue Sep 8 11:52:31 2020
hgvs: runtime 1330.173s
dbsnp: finished at Tue Sep 8 11:53:55 2020
dbsnp: runtime 1125.568s
annotator(s) finished in 1416.194s
Running aggregator...
Variants Traceback (most recent call last):
File "/data/umcg-mterpstra/apps/software/open-cravat/1.8.0-foss-2018a-Python-3.6.4/lib/python3.6/site-packages/cravat/cravat_class.py", line 382, in main
self.result_path = self.run_aggregator()
File "/data/umcg-mterpstra/apps/software/open-cravat/1.8.0-foss-2018a-Python-3.6.4/lib/python3.6/site-packages/cravat/cravat_class.py", line 905, in run_aggregator
v_aggregator.run()
File "/data/umcg-mterpstra/apps/software/open-cravat/1.8.0-foss-2018a-Python-3.6.4/lib/python3.6/site-packages/cravat/aggregator.py", line 141, in run
for lnum, line, rd in reader.loop_data():
File "/data/umcg-mterpstra/apps/software/open-cravat/1.8.0-foss-2018a-Python-3.6.4/lib/python3.6/site-packages/cravat/inout.py", line 155, in loop_data
tok = json.loads(tok)
File "/software/software/Python/3.6.4-foss-2018a/lib/python3.6/json/__init__.py", line 354, in loads
return _default_decoder.decode(s)
File "/software/software/Python/3.6.4-foss-2018a/lib/python3.6/json/decoder.py", line 339, in decode
obj, end = self.raw_decode(s, idx=_w(s, 0).end())
File "/software/software/Python/3.6.4-foss-2018a/lib/python3.6/json/decoder.py", line 357, in raw_decode
raise JSONDecodeError("Expecting value", s, err.value) from None
json.decoder.JSONDecodeError: Expecting value: line 1 column 1 (char 0)
It should be relatively straightforward to write the tsv to a tsv.gz via a pipe, and often there is a performance gain by reducing the amount of disk IO.
Is this feature compelling or undesirable?
Hi!
Is there a way to add a column, on gene view table, with the number of samples (sample_count) that have variants in a particular gene? Right now this information is only available in the variant table, but for each individual variant.
The vcf reporter currently outputs one field (CRV) when pipe (|) delimited values. It would be easier to work with the vcf if each field was separate. For example, there is no way generate a nice report using https://github.com/igvteam/igv-reports with the current vcf output. Thanks.
Hi! It's not a bug report, just a suggestion:
I've noticed that the content of the "secondary_data" variable varies from annotator to annotator when the fields are empty.
Here is an example of rare variant, which absent from the frequencies DBs:
print(secondary_data['esp6500'])
[]
print(secondary_data['gnomad3'])
[{'uid': 1, 'af': None, 'af_afr': None, 'af_asj': None, 'af_eas': None, 'af_fin': None, 'af_lat': None, 'af_nfe': None, 'af_oth': None, 'af_sas': None}]
print(secondary_data['thousandgenomes'])
[{'uid': 1, 'af': None, 'afr_af': None, 'amr_af': None, 'eas_af': None, 'eur_af': None, 'sas_af': None}]
For example clinvar also behave similar to esp6500
This is quite inconvenient to handle and also there is no guarantee that the output format will not change back and forth.
So It would be awesome to have some convention either to return empty list or dict with Nones (the later makes more sense probably)
And I suppose it's couple lines of the code
Best, Eugene
I've had issues getting the gui/webserver to work so I switched to just going straight command line. As a matter of best practices I setup a new virtual environment(python 3.7.4 and latest pip version) for a fresh install of open cravat. When I try to run:
cravat-admin install-base
After installing open cravat via pip I'm getting this output:
Traceback (most recent call last): File "/home/paul/Work/opencravat/ocrav/bin/cravat-admin", line 8, in <module> sys.exit(main()) File "/home/paul/Work/opencravat/ocrav/lib/python3.7/site-packages/cravat/cravat_admin.py", line 693, in main args.func(args) File "/home/paul/Work/opencravat/ocrav/lib/python3.7/site-packages/cravat/cravat_admin.py", line 393, in install_base install_modules(args) File "/home/paul/Work/opencravat/ocrav/lib/python3.7/site-packages/cravat/cravat_admin.py", line 277, in install_modules matching_names = au.search_remote(*args.modules) File "/home/paul/Work/opencravat/ocrav/lib/python3.7/site-packages/cravat/admin_util.py", line 378, in search_remote for module_name in list_remote(): File "/home/paul/Work/opencravat/ocrav/lib/python3.7/site-packages/cravat/admin_util.py", line 356, in list_remote return sorted(list(mic.remote.keys())) AttributeError: 'str' object has no attribute 'keys'
I'am using the code script: oc module install-base , how the connection to karchinlab.org was disrupted.
WARNING: Could not list modules from http://karchinlab.org/cravatstore/manifest.yml. Check internet connection.
No modules to install found
Hi,
When i'm using a VPN and try to access to my openCravat server, i'm getting a "Lost connection to server\nPlease launch OpenCRAVAT again" popup. This error manifests itself in 2 ways :
Remarks :
ping on the webserver show 13ms of latency and no packet lost :
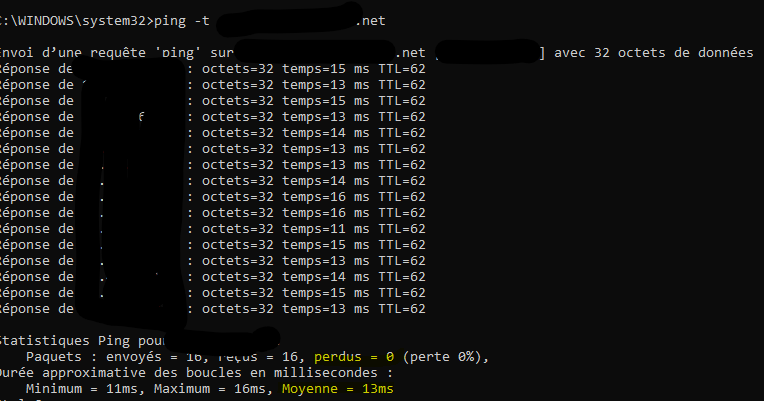
Is there a way to increase timeout for heartbeat ?
Let me know if I can add/do some test.
Thank you for your help.
test
Is it possible to load automatically a corresponding BAM file with IGV widget in order to visualize the variant ? Instead of loading it manually each time I open an VCF.
is it a subroutine of text or excel perhaps?
Hi
I am running your tool on a server, and I cannot figure out which port it is using?
Please advise
I have another app which also requires 8080. As a result, this has to be closed before running openCRAVAT. Is there a way to change the port used by openCRAVAT?
Many thanks for your help
test1.vcf
##fileformat=VCFv4.2
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT test1
chr11 113400106 . G A 1339.06 PASS AC=2;AF=1;AN=2;DP=432;FS=0;MQ=249.72;QD=3.1;SOR=0.94;FractionInformativeReads=1;VQSLOD=5.35516 GT:AD:AF:DP:F1R2:F2R1:GQ:PL:GP:PRI:SB:MB 1/1:0,432:1:432:0,246:0,186:450:1377,1292,0:450,450,0:0,34.77,37.77:0,0,192,240:0,0,212,220
chr11 113412966 . C A 224.21 PASS AC=2;AF=1;AN=2;DP=59;FS=0;MQ=250;QD=3.8;SOR=2.507;FractionInformativeReads=1;VQSLOD=7.44863 GT:AD:AF:DP:F1R2:F2R1:GQ:PL:GP:PRI:SB:MB 1/1:0,59:1:59:0,26:0,33:174:262,177,0:224.21,174.21,0:0,34.77,37.77:0,0,13,46:0,0,32,27
command line
oc run test1.vcf --liftover hg38 -x --cleanrun -t text -d out-test1 -a pharmgkb
The first record has no identified pharmgkb match, but should match
https://www.pharmgkb.org/variant/PA166154339
The second record does correctly have a pharmgkb match
https://www.pharmgkb.org/variant/PA166154313
can anyone explain why open-cravat does not identify PA166154339?
oc run test.vcf.gz --repeat converter -t vcf -l hg38 -d .
File "/opt/anaconda3/envs/cravat/lib/python3.8/site-packages/cravat/cravat_report.py", line 240, in run_level
self.write_preface(level)
File "/Users/Shared/open-cravat/modules/reporters/vcfreporter/vcfreporter.py", line 91, in write_preface
for line in f:
File "/opt/anaconda3/envs/cravat/lib/python3.8/codecs.py", line 322, in decode
(result, consumed) = self._buffer_decode(data, self.errors, final)
UnicodeDecodeError: 'utf-8' codec can't decode byte 0x8b in position 1: invalid start byte
Could you update cosmic to the newest v91 version?
I'm having an issue where I get empty results when I try to do a fairly standard annotation run. Here's the command I've used:
cravat my_data.vcf -l hg38 -t tsv
Here's the output:
Input file(s): my_data.vcf
Genome assembly: hg38
Running converter...
Converter (converter) finished in 2.788s
Running gene mapper...
UCSC hg38 Gene Mapper (hg38) finished in 0.038s
Running annotators...
revel: started at Wed Jan 22 08:03:15 2020
revel: finished at Wed Jan 22 08:03:15 2020
revel: runtime 0.001s
hgvs: started at Wed Jan 22 08:03:15 2020
hgvs: finished at Wed Jan 22 08:03:15 2020
hgvs: runtime 0.002s
clinvar: started at Wed Jan 22 08:03:15 2020
sift: started at Wed Jan 22 08:03:15 2020
phylop: started at Wed Jan 22 08:03:15 2020
clinvar: finished at Wed Jan 22 08:03:15 2020
clinvar: runtime 0.002s
sift: finished at Wed Jan 22 08:03:15 2020
sift: runtime 0.001s
gnomad: started at Wed Jan 22 08:03:15 2020
phylop: finished at Wed Jan 22 08:03:15 2020
phylop: runtime 0.002s
vest: started at Wed Jan 22 08:03:15 2020
gnomad: finished at Wed Jan 22 08:03:15 2020
gnomad: runtime 0.002s
vest: finished at Wed Jan 22 08:03:15 2020
vest: runtime 0.009s
polyphen2: started at Wed Jan 22 08:03:15 2020
polyphen2: finished at Wed Jan 22 08:03:15 2020
polyphen2: runtime 0.001s
annotator(s) finished in 1.147s
Running aggregator...
Variants finished in 1.054s
Genes finished in 0.923s
Samples finished in 1.094s
Tags finished in 2.050s
Running postaggregators...
VCF Info (vcfinfo) finished in 0.430s
Tag Sampler (tagsampler) finished in 0.573s
Running reporter...
TSV Reporter (tsvreporter)
hg38: started getting gene summary data
hg38: finished getting gene summary data in 0.000s
vest: getting gene summary data
vest: finished getting gene summary data in 0.000s
finished in 0.296s
Finished normally. Runtime: 10.741s
Here's an example row of the data (I've changed/spoofed some of the values):
chr3 | 7966478 | . | A | G | 44 | PASS | P=0.999544 | GT:RV:VV:ZQ:GQ:PL | 1/1:30:0:20:.:1228,217,0
$ cravat-admin install-base
Finished installation of excelreporter:1.0.2
Traceback (most recent call last):
File "/usr/local/lib/python3.7/site-packages/cravat/admin_util.py", line 610, in install_module
install_module('wg' + module_name)
File "/usr/local/lib/python3.7/site-packages/cravat/admin_util.py", line 534, in install_module
version = get_remote_latest_version(module_name)
File "/usr/local/lib/python3.7/site-packages/cravat/admin_util.py", line 394, in get_remote_latest_version
return mic.remote[module_name]['latest_version']
KeyError: 'wgexcelreporter'
Sorry if it is written somewhere, but I failed to find it.
I need to build an annotator, which takes into account results from several other annotators. As fas as I get it - the option seondary_data designed exactly for that purpouse. But I have not found a clear doc on how to use it. I might figure it out from the code, but it'll take some time for me, so I appreciate if you can point me out where to look for the right info. Also I believe that if it's not in the docs now it anyhow should be there.
Best wishes, Eugene
Example: distributed 1.22.0 requires msgpack, which is not installed.
tensorboard 1.8.0 has requirement bleach==1.5.0, but you'll have bleach 2.1.3 which is incompatible.
tensorboard 1.8.0 has requirement html5lib==0.9999999, but you'll have html5lib 1.0.1 which is incompatible.
I'm trying to install open-cravat for our team with a shared resource. I do not have sudo privileges. My command line, as follows, attempts to place the product into our space.
pip3 install
--verbose
--install-option="--prefix=/group3/tools"
open-cravat
....
Running command /usr/bin/python3 -u -c "import setuptools, tokenize;__file__='/tmp/pip-build-w_xiajc1/pyyaml/setup.py'; \
f=getattr(tokenize, 'open', open)(__file__);code=f.read().replace('\r\n', '\n');\
f.close();exec(compile(code, __file__, 'exec'))" install --record /tmp/pip-tfeikwwc-record/install-record.txt \
--single-version-externally-managed --compile --prefix=/group3/tools
....
copying lib3/yaml/scanner.py -> build/lib.linux-x86_64-3.6/yaml
running build_ext
creating build/temp.linux-x86_64-3.6
checking if libyaml is compilable
gcc -pthread -Wno-unused-result -Wsign-compare -DNDEBUG -O2 -g -pipe -Wall -Wp,-D_FORTIFY_SOURCE=2 -fexceptions -fstack-protector-strong $
checking if libyaml is linkable
gcc -pthread build/temp.linux-x86_64-3.6/check_libyaml.o -L/usr/lib64 -lyaml -o build/temp.linux-x86_64-3.6/check_libyaml
building '_yaml' extension
creating build/temp.linux-x86_64-3.6/ext
gcc -pthread -Wno-unused-result -Wsign-compare -DNDEBUG -O2 -g -pipe -Wall -Wp,-D_FORTIFY_SOURCE=2 -fexceptions -fstack-protector-strong $
ext/_yaml.c:4:20: fatal error: Python.h: No such file or directory
#include "Python.h"
^
compilation terminated.
error: command 'gcc' failed with exit status 1
Running setup.py install for pyyaml: finished with status 'error'
Adding -v to the last action ("Running command") I see
import 'setuptools.msvc' # <_frozen_importlib_external.SourceFileLoader object at 0x2ae8d794b5c0>
import 'setuptools' # <_frozen_importlib_external.SourceFileLoader object at 0x2ae8cac41a20>
Traceback (most recent call last):
File "", line 1, in
File "/usr/lib64/python3.6/tokenize.py", line 452, in open buffer = _builtin_open(filename, 'rb')
FileNotFoundError: [Errno 2] No such file or directory: '/tmp/pip-build-ydzqg6mr/pyyaml/setup.py'
I was wondering if there was a connect between "msvc" and not find "Python.h" in a case insensitive manor?
Any help very much appreciated.
I don't know when to reinstall/update opencravat because its not easy to find a chronology of all the new features coming out at lightning speed!
The query
select clinvar__disease_names from variant where clinvar__id = 184594;
returns
Hereditary cancer-predisposing syndrome|Neurofibromatosis, type 1|Café-au-lait macules with pulmonary stenosis|Neurofibromatosis, familial spinal|Neurofibromatosis-Noonan syndrome|not specified|not provided
which is broadly consistent with
https://www.ncbi.nlm.nih.gov/clinvar/variation/184594/
however the unicode on Café-au-lait seems to have been mangled. If there is a correct way I should be decoding that please let me know, but as is, I suspect a unicode issue on whatever code was importing from clinvar.
Hello!
Thanks for creating and maintaining open-cravat - such a great annotation tool!
I am annotating with:
cravat \
sample.tsv \
-n $bname \
-a chasmplus chasmplus_BRCA polyphen2 fathmm cosmic cosmic_gene cancer_hotspots \
-d sample_out -l hg38 \
--mp 10 \
-t text
and I'm getting all the annotations but cancer_hotspots: it returned NA in all mutations for all samples.
(I have installed the cancer_hotstpots annotator).
I realize that the original database https://www.cancerhotspots.org/#/home
is along hg19/b37: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5808190/,
but I was hoping that open-cravat does the liftover.
Has anybody tacked this issue before?
Thanks!
Sergey
Hello,
Thank you for this great tool.
I installed openCravat on my linux server and it run well.
But I have issue to run multiuser mode.
I installed opencravat and open-cravat-multiuser with pip/conda/docker but it is the same issue.
When I ran oc gui --multiuser the openCravat web page (0.0.0.0:8080) isn't avaible.
For example for docker installation:
I followed docker hub instruction for linux:
In the docker-compose.yml I changed this : command: ['oc', 'gui'] to command: ['oc', 'gui' , '--multiuser' ] for run multiuser mode.
But when I run the docker with : docker-compose up -d
the web page (0.0.0.0:8080) it'not avaible.
So I was woddering if I did somthing wrong?
Or if I have to first setup in particular way my server before make the openCravat installation?
Thank you again.
Ivaylo
Hi, I've noticed some strange thing in the liftover results
my input vcf file has following line:
chr10 48414222 rs556004917 G A
For reasons unknow, with the command oc run test.vcf -l hg19 --skip reporter -a dbsnp I'm getting the output with hg38 coordinates: 47325138 (see the attach figure)
But it has to be 47325140 (even alt/ref now incorrect!) ! I'm not sure what is going on here and would appriciate any advice!
PS UCSC liftover tools (https://genome.ucsc.edu/cgi-bin/hgLiftOver) also convert 48414222->47325140
Best, Eugene
conda create --name cravat
conda activate cravat
conda install pip
pip install open-cravat
oc module install --yes clinvar vcfreporter vcf-converter hg38
wget https://raw.githubusercontent.com/vcflib/vcflib/master/samples/sample.vcf
oc run sample.vcf -t vcf -l hg38
Hi!
I tried to download a filtered table (only chr X variants) as an excel file but there is no option. It exports it as tsv. I can only export the whole table as an excel file. The problem is that having many annotators makes this big excel file unusable.
Is there a way to do that that I have not noticed?
Hi!
What is recommended solution to run opencravat multiuser server on Windows Server 2016 startup/background without user logging in (as a service)?
Error when an header ID contains non-alphanumeric char (like + or . )
Example:
#INFO=<ID=gnomad.exome,Number=.,Type=String,Description="/GNOMAD/2.1.1/gnomad.exomes.r2.1.1.sites.vcf.gz (exact)">
Error:
Traceback (most recent call last):
File "/usr/local/lib/python3.6/site-packages/cravat/cravat_class.py", line 382, in main
self.result_path = self.run_aggregator()
File "/usr/local/lib/python3.6/site-packages/cravat/cravat_class.py", line 905, in run_aggregator
v_aggregator.run()
File "/usr/local/lib/python3.6/site-packages/cravat/aggregator.py", line 94, in run
self._setup()
File "/usr/local/lib/python3.6/site-packages/cravat/aggregator.py", line 294, in _setup
self._setup_table()
File "/usr/local/lib/python3.6/site-packages/cravat/aggregator.py", line 351, in _setup_table
self.cursor.execute(q)
sqlite3.OperationalError: near ".": syntax error
A declarative, efficient, and flexible JavaScript library for building user interfaces.
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
An Open Source Machine Learning Framework for Everyone
The Web framework for perfectionists with deadlines.
A PHP framework for web artisans
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
Some thing interesting about web. New door for the world.
A server is a program made to process requests and deliver data to clients.
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
Some thing interesting about visualization, use data art
Some thing interesting about game, make everyone happy.
We are working to build community through open source technology. NB: members must have two-factor auth.
Open source projects and samples from Microsoft.
Google ❤️ Open Source for everyone.
Alibaba Open Source for everyone
Data-Driven Documents codes.
China tencent open source team.