Comments (11)
Hi Eugene, thanks for letting us know. We'll take a look.
from open-cravat.
@EugeneEA The issue was due to how OpenCRAVAT was handling liftover for the regions which were inverted between hg19 and hg38. This fix will be included in our next release of OpenCRAVAT. If you want to try this fix now, a workaround is:
1. Find and download cravat_convert.py in 2.1.1 branch of this repo.
2. Put the cravat_convert.py into your OpenCRAVAT pip package directory (which can be found with "pip list -v|grep open-cravat". Add "cravat" to the found site-packages directory).
from open-cravat.
Awesome - thanks!
PS I was suspecting that it is due to complicated changes in that region between builds, but I thought that under the hood Cravat uses some third party software for lift over task.
Best, Eugene
from open-cravat.
@EugeneEA Yes, OpenCRAVAT uses pyliftover internally and pyliftover uses 0-based coordinates while OpenCRAVAT uses 1-based coordinates. How OpenCRAVAT handled the communication between the two was the cause of the issue related to the genomic regions which were inverted from hg19 to hg38.
from open-cravat.
Hi, I'veagain noticed some problems with liftover procedure:
here is the results of hg19 mapped vcf, submitted to the oc (version 2.2.3) with -l hg19 parameter, notice the absence of the gnomad3 annotation:

and here is the same vcf file, which was lifted over to hg38 with gatk picard tool, as you can see now, the gnomad3 annotation is present:
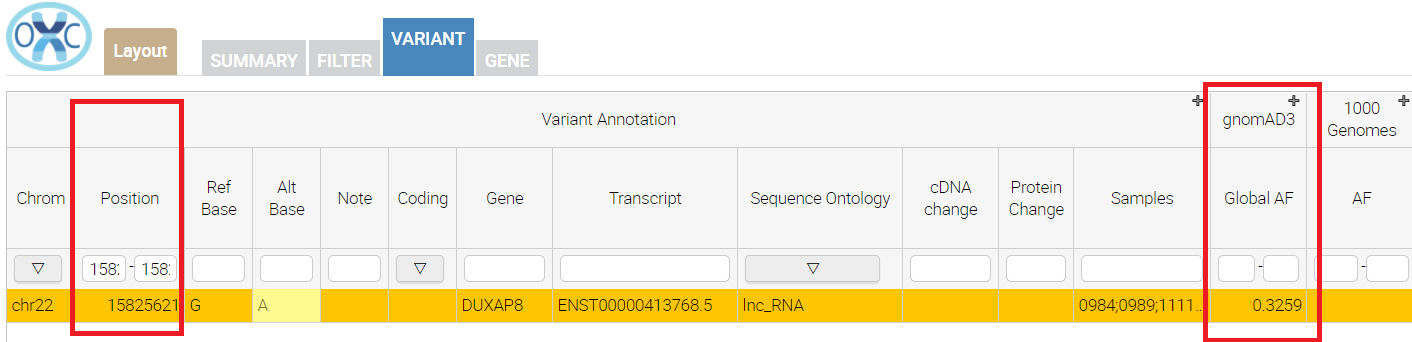
Basically cravat maneges to liftover the position but failed to swap alleles (the ref allele was changed between hg19 to hg38), therefore new SNP became unic in the hg38 and absent from gnomad. Could you, please, comment on this bug? Should I just routinely run picard before cravat? (btw why you do not use pickard internally?)
Best, Eugene
from open-cravat.
@EugeneEA Thanks for finding the bug and sorry for the inconvenience. A fix will be included in the next release of OpenCRAVAT. Meanwhile, you can try the fix as follows.
- Download cravat_convert.py at
https://drive.google.com/file/d/1y5Yx1tiJoM5jd380bTiWIQLkYOWCDeas/view?usp=sharing pip list -v|grep open-cravat
(This will show you where OpenCRAVAT is installed.)- Overwrite cravat_convert.py in the folder found in (2) with the downloaded cravat_convert.py.
Let me know if works for you.
We didn't use Picard because we wanted to keep minimal dependencies and to use only Python dependencies.
from open-cravat.
@rkimoakbioinformatics thanks a lot for such quick reply! It did work out,
best, Eugene
from open-cravat.
@EugeneEA That's great. Glad it worked.
from open-cravat.
@rkimoakbioinformatics Hi, I've noticed that you have not change the file in your latest version? was it by mistake or are there any meaningfull reasons?
Best, Eugene
from open-cravat.
@EugeneEA If you mean the fix at #35 (comment), this fix came after the latest version, open-cravat 2.2.3. We are close to releasing the next version, open-cravat 2.2.4, and this version will have the fixed cravat_convert.py file.
from open-cravat.
@EugeneEA I hope that OpenCRAVAT is working for you well. We are conducting interviews to learn what else we could do for genomic variant researchers. Would you be available for such an interview? It will take just about 15 minutes and your input will be invaluable to make OpenCRAVAT even better. It does not have to be about your experience with OpenCRAVAT. We just would like to hear what your experience has been with genomic variant analysis tools. If so, please let me know at [email protected].
from open-cravat.
Related Issues (20)
- UniProt HOT 2
- Error in IGV widget HOT 1
- pharmgkb OC and website differing annotations HOT 7
- gnomad update HOT 1
- Variant Conversion HOT 2
- rdata download option not working HOT 1
- silent failures from multiple annotators HOT 2
- Support for Spanning or overlapping deletions HOT 1
- Access to error logs from web server HOT 1
- Homozygous reference variants HOT 1
- KeyError in sample_id with multiple VCF files HOT 1
- New databases creation HOT 4
- Single variant tab not working! HOT 2
- python 3.12 module imp deprecated HOT 1
- Ambiguous annotation results HOT 1
- incorrect SO assignment - "exon_loss_variant" for intronic region
- IndexError: index out of range when annotating vcf file HOT 1
- How to update Open-CRAVAT from v2.2.9 to v2.5.0 ? HOT 1
- Update link in OC versions <v2.5 is unreadable and unusable HOT 1
- oc module install-base issue HOT 2
Recommend Projects
-
 React
React
A declarative, efficient, and flexible JavaScript library for building user interfaces.
-
Vue.js
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
-
 Typescript
Typescript
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
-
TensorFlow
An Open Source Machine Learning Framework for Everyone
-
Django
The Web framework for perfectionists with deadlines.
-
Laravel
A PHP framework for web artisans
-
D3
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
-
Recommend Topics
-
javascript
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
-
web
Some thing interesting about web. New door for the world.
-
server
A server is a program made to process requests and deliver data to clients.
-
Machine learning
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
-
Visualization
Some thing interesting about visualization, use data art
-
Game
Some thing interesting about game, make everyone happy.
Recommend Org
-
Facebook
We are working to build community through open source technology. NB: members must have two-factor auth.
-
Microsoft
Open source projects and samples from Microsoft.
-
Google
Google ❤️ Open Source for everyone.
-
Alibaba
Alibaba Open Source for everyone
-
D3
Data-Driven Documents codes.
-
Tencent
China tencent open source team.

from open-cravat.