MUSCLE is widely-used software for making multiple alignments of biological sequences.
Version 5 of MUSCLE achieves highest scores on Balibase, Bralibase and Balifam benchmark tests and scales to thousands of sequences on a commodity desktop computer.
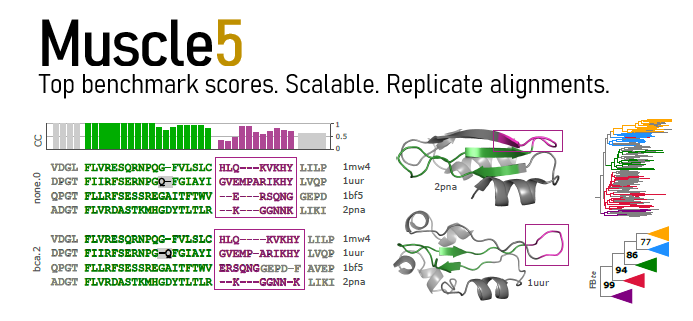
This version supports generating an ensemble of alternative alignments with the same high accuracy obtained with default parameters. By comparing downstream predictions from different alignments, such as trees, a biologist can evaluation the robustness of conclusions against alignment errors.
Binary files are self-contained, no dependencies.
https://github.com/rcedgar/muscle/releases
https://github.com/rcedgar/muscle/wiki/Building-MUSCLE
Edgar, Robert C. Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nature Communications 13.1 (2022): 6968.
https://www.nature.com/articles/s41467-022-34630-w.pdf