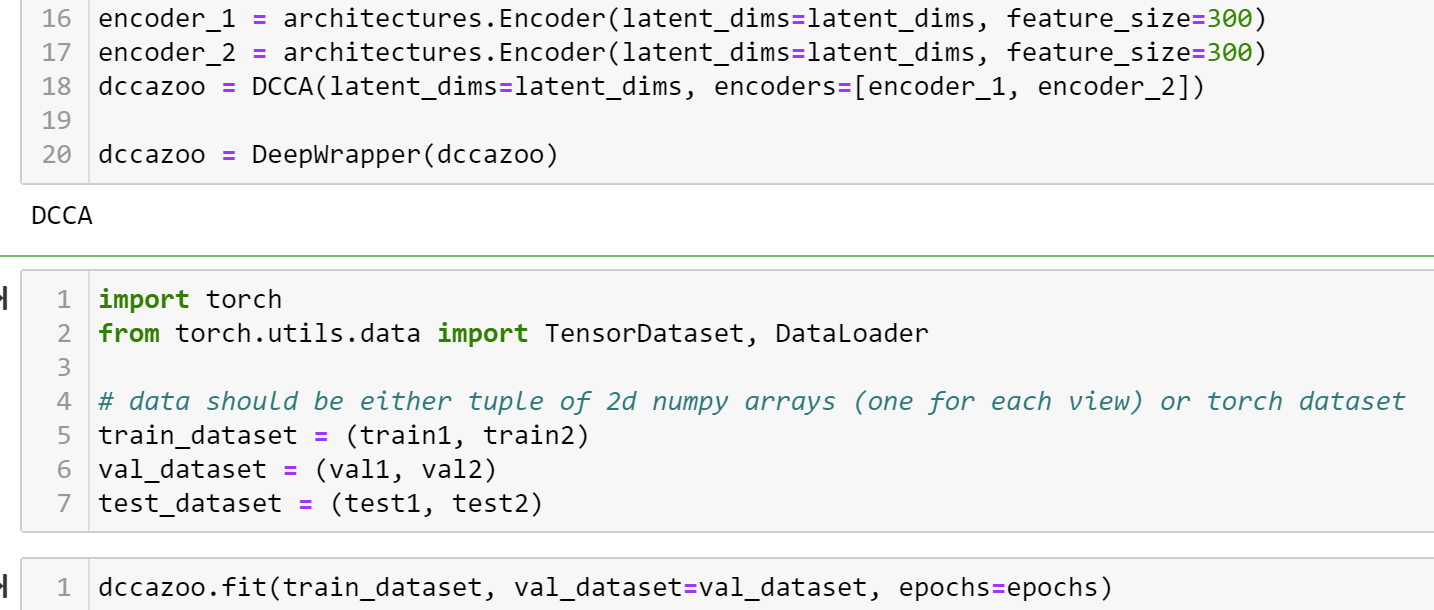
Does the DCCA model not support float values? Because I get the error below when I try to train the model.
I got the code working before the shift to pylightning. My data looks like this

RuntimeError Traceback (most recent call last)
/tmp/ipykernel_22306/563663734.py in <module>
12
13 trainer = pl.Trainer(max_epochs=epochs, enable_checkpointing=False)
---> 14 trainer.fit(dcca.float(), train_loader, val_loader)
15 print("Time taken to train:", datetime.now() - then)
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py in fit(self, model, train_dataloaders, val_dataloaders, datamodule, train_dataloader, ckpt_path)
735 )
736 train_dataloaders = train_dataloader
--> 737 self._call_and_handle_interrupt(
738 self._fit_impl, model, train_dataloaders, val_dataloaders, datamodule, ckpt_path
739 )
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py in _call_and_handle_interrupt(self, trainer_fn, *args, **kwargs)
680 """
681 try:
--> 682 return trainer_fn(*args, **kwargs)
683 # TODO: treat KeyboardInterrupt as BaseException (delete the code below) in v1.7
684 except KeyboardInterrupt as exception:
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py in _fit_impl(self, model, train_dataloaders, val_dataloaders, datamodule, ckpt_path)
770 # TODO: ckpt_path only in v1.7
771 ckpt_path = ckpt_path or self.resume_from_checkpoint
--> 772 self._run(model, ckpt_path=ckpt_path)
773
774 assert self.state.stopped
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py in _run(self, model, ckpt_path)
1193
1194 # dispatch `start_training` or `start_evaluating` or `start_predicting`
-> 1195 self._dispatch()
1196
1197 # plugin will finalized fitting (e.g. ddp_spawn will load trained model)
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py in _dispatch(self)
1273 self.training_type_plugin.start_predicting(self)
1274 else:
-> 1275 self.training_type_plugin.start_training(self)
1276
1277 def run_stage(self):
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/plugins/training_type/training_type_plugin.py in start_training(self, trainer)
200 def start_training(self, trainer: "pl.Trainer") -> None:
201 # double dispatch to initiate the training loop
--> 202 self._results = trainer.run_stage()
203
204 def start_evaluating(self, trainer: "pl.Trainer") -> None:
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py in run_stage(self)
1283 if self.predicting:
1284 return self._run_predict()
-> 1285 return self._run_train()
1286
1287 def _pre_training_routine(self):
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py in _run_train(self)
1305 self.progress_bar_callback.disable()
1306
-> 1307 self._run_sanity_check(self.lightning_module)
1308
1309 # enable train mode
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py in _run_sanity_check(self, ref_model)
1369 # run eval step
1370 with torch.no_grad():
-> 1371 self._evaluation_loop.run()
1372
1373 self.call_hook("on_sanity_check_end")
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/loops/base.py in run(self, *args, **kwargs)
143 try:
144 self.on_advance_start(*args, **kwargs)
--> 145 self.advance(*args, **kwargs)
146 self.on_advance_end()
147 self.restarting = False
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/loops/dataloader/evaluation_loop.py in advance(self, *args, **kwargs)
108 dl_max_batches = self._max_batches[dataloader_idx]
109
--> 110 dl_outputs = self.epoch_loop.run(dataloader, dataloader_idx, dl_max_batches, self.num_dataloaders)
111
112 # store batch level output per dataloader
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/loops/base.py in run(self, *args, **kwargs)
143 try:
144 self.on_advance_start(*args, **kwargs)
--> 145 self.advance(*args, **kwargs)
146 self.on_advance_end()
147 self.restarting = False
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/loops/epoch/evaluation_epoch_loop.py in advance(self, data_fetcher, dataloader_idx, dl_max_batches, num_dataloaders)
120 # lightning module methods
121 with self.trainer.profiler.profile("evaluation_step_and_end"):
--> 122 output = self._evaluation_step(batch, batch_idx, dataloader_idx)
123 output = self._evaluation_step_end(output)
124
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/loops/epoch/evaluation_epoch_loop.py in _evaluation_step(self, batch, batch_idx, dataloader_idx)
215 self.trainer.lightning_module._current_fx_name = "validation_step"
216 with self.trainer.profiler.profile("validation_step"):
--> 217 output = self.trainer.accelerator.validation_step(step_kwargs)
218
219 return output
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/accelerators/accelerator.py in validation_step(self, step_kwargs)
234 """
235 with self.precision_plugin.val_step_context():
--> 236 return self.training_type_plugin.validation_step(*step_kwargs.values())
237
238 def test_step(self, step_kwargs: Dict[str, Union[Any, int]]) -> Optional[STEP_OUTPUT]:
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/pytorch_lightning/plugins/training_type/training_type_plugin.py in validation_step(self, *args, **kwargs)
217
218 def validation_step(self, *args, **kwargs):
--> 219 return self.model.validation_step(*args, **kwargs)
220
221 def test_step(self, *args, **kwargs):
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/cca_zoo/deepmodels/trainers.py in validation_step(self, batch, batch_idx)
55 def validation_step(self, batch, batch_idx):
56 data, label = batch
---> 57 loss = self.model.loss(*data)
58 self.log("val loss", loss)
59 return loss
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/cca_zoo/deepmodels/dcca.py in loss(self, *args)
53 :return:
54 """
---> 55 z = self(*args)
56 return self.objective.loss(*z)
57
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/torch/nn/modules/module.py in _call_impl(self, *input, **kwargs)
1100 if not (self._backward_hooks or self._forward_hooks or self._forward_pre_hooks or _global_backward_hooks
1101 or _global_forward_hooks or _global_forward_pre_hooks):
-> 1102 return forward_call(*input, **kwargs)
1103 # Do not call functions when jit is used
1104 full_backward_hooks, non_full_backward_hooks = [], []
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/cca_zoo/deepmodels/dcca.py in forward(self, *args)
43 z = []
44 for i, encoder in enumerate(self.encoders):
---> 45 z.append(encoder(args[i]))
46 return z
47
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/torch/nn/modules/module.py in _call_impl(self, *input, **kwargs)
1100 if not (self._backward_hooks or self._forward_hooks or self._forward_pre_hooks or _global_backward_hooks
1101 or _global_forward_hooks or _global_forward_pre_hooks):
-> 1102 return forward_call(*input, **kwargs)
1103 # Do not call functions when jit is used
1104 full_backward_hooks, non_full_backward_hooks = [], []
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/cca_zoo/deepmodels/architectures.py in forward(self, x)
66
67 def forward(self, x):
---> 68 x = self.layers(x)
69 if self.variational:
70 mu = self.fc_mu(x)
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/torch/nn/modules/module.py in _call_impl(self, *input, **kwargs)
1100 if not (self._backward_hooks or self._forward_hooks or self._forward_pre_hooks or _global_backward_hooks
1101 or _global_forward_hooks or _global_forward_pre_hooks):
-> 1102 return forward_call(*input, **kwargs)
1103 # Do not call functions when jit is used
1104 full_backward_hooks, non_full_backward_hooks = [], []
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/torch/nn/modules/container.py in forward(self, input)
139 def forward(self, input):
140 for module in self:
--> 141 input = module(input)
142 return input
143
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/torch/nn/modules/module.py in _call_impl(self, *input, **kwargs)
1100 if not (self._backward_hooks or self._forward_hooks or self._forward_pre_hooks or _global_backward_hooks
1101 or _global_forward_hooks or _global_forward_pre_hooks):
-> 1102 return forward_call(*input, **kwargs)
1103 # Do not call functions when jit is used
1104 full_backward_hooks, non_full_backward_hooks = [], []
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/torch/nn/modules/container.py in forward(self, input)
139 def forward(self, input):
140 for module in self:
--> 141 input = module(input)
142 return input
143
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/torch/nn/modules/module.py in _call_impl(self, *input, **kwargs)
1100 if not (self._backward_hooks or self._forward_hooks or self._forward_pre_hooks or _global_backward_hooks
1101 or _global_forward_hooks or _global_forward_pre_hooks):
-> 1102 return forward_call(*input, **kwargs)
1103 # Do not call functions when jit is used
1104 full_backward_hooks, non_full_backward_hooks = [], []
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/torch/nn/modules/linear.py in forward(self, input)
101
102 def forward(self, input: Tensor) -> Tensor:
--> 103 return F.linear(input, self.weight, self.bias)
104
105 def extra_repr(self) -> str:
~/nobackup/miniconda3/envs/matchms/lib/python3.8/site-packages/torch/nn/functional.py in linear(input, weight, bias)
1846 if has_torch_function_variadic(input, weight, bias):
1847 return handle_torch_function(linear, (input, weight, bias), input, weight, bias=bias)
-> 1848 return torch._C._nn.linear(input, weight, bias)
1849
1850
RuntimeError: expected scalar type Double but found Float```