🐳 Paper is now available on arXiv!
Simple and Practical Python library for CMA-ES. Please refer to the paper [Nomura and Shibata 2024] for detailed information, including the design philosophy and advanced examples.
Supported Python versions are 3.7 or later.
$ pip install cmaes
Or you can install via conda-forge.
$ conda install -c conda-forge cmaes
This library provides an "ask-and-tell" style interface. We employ the standard version of CMA-ES [Hansen 2016].
import numpy as np
from cmaes import CMA
def quadratic(x1, x2):
return (x1 - 3) ** 2 + (10 * (x2 + 2)) ** 2
if __name__ == "__main__":
optimizer = CMA(mean=np.zeros(2), sigma=1.3)
for generation in range(50):
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = quadratic(x[0], x[1])
solutions.append((x, value))
print(f"#{generation} {value} (x1={x[0]}, x2 = {x[1]})")
optimizer.tell(solutions)And you can use this library via Optuna [Akiba et al. 2019], an automatic hyperparameter optimization framework. Optuna's built-in CMA-ES sampler which uses this library under the hood is available from v1.3.0 and stabled at v2.0.0. See the documentation or v2.0 release blog for more details.
import optuna
def objective(trial: optuna.Trial):
x1 = trial.suggest_uniform("x1", -4, 4)
x2 = trial.suggest_uniform("x2", -4, 4)
return (x1 - 3) ** 2 + (10 * (x2 + 2)) ** 2
if __name__ == "__main__":
sampler = optuna.samplers.CmaEsSampler()
study = optuna.create_study(sampler=sampler)
study.optimize(objective, n_trials=250)The performance of the CMA-ES can deteriorate when faced with difficult problems such as multimodal or noisy ones, if its hyperparameter values are not properly configured. The Learning Rate Adaptation CMA-ES (LRA-CMA) effectively addresses this issue by autonomously adjusting the learning rate. Consequently, LRA-CMA eliminates the need for expensive hyperparameter tuning.
LRA-CMA can be used by simply adding lr_adapt=True to the initialization of CMA().
Source code
import numpy as np
from cmaes import CMA
def rastrigin(x):
dim = len(x)
return 10 * dim + sum(x**2 - 10 * np.cos(2 * np.pi * x))
if __name__ == "__main__":
dim = 40
optimizer = CMA(mean=3*np.ones(dim), sigma=2.0, lr_adapt=True)
for generation in range(50000):
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = rastrigin(x)
if generation % 500 == 0:
print(f"#{generation} {value}")
solutions.append((x, value))
optimizer.tell(solutions)
if optimizer.should_stop():
breakThe full source code is available here.
Warm Starting CMA-ES (WS-CMA) is a method that transfers prior knowledge from similar tasks through the initialization of the CMA-ES. This is useful especially when the evaluation budget is limited (e.g., hyperparameter optimization of machine learning algorithms).
Source code
import numpy as np
from cmaes import CMA, get_warm_start_mgd
def source_task(x1: float, x2: float) -> float:
b = 0.4
return (x1 - b) ** 2 + (x2 - b) ** 2
def target_task(x1: float, x2: float) -> float:
b = 0.6
return (x1 - b) ** 2 + (x2 - b) ** 2
if __name__ == "__main__":
# Generate solutions from a source task
source_solutions = []
for _ in range(1000):
x = np.random.random(2)
value = source_task(x[0], x[1])
source_solutions.append((x, value))
# Estimate a promising distribution of the source task,
# then generate parameters of the multivariate gaussian distribution.
ws_mean, ws_sigma, ws_cov = get_warm_start_mgd(
source_solutions, gamma=0.1, alpha=0.1
)
optimizer = CMA(mean=ws_mean, sigma=ws_sigma, cov=ws_cov)
# Run WS-CMA-ES
print(" g f(x1,x2) x1 x2 ")
print("=== ========== ====== ======")
while True:
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = target_task(x[0], x[1])
solutions.append((x, value))
print(
f"{optimizer.generation:3d} {value:10.5f}"
f" {x[0]:6.2f} {x[1]:6.2f}"
)
optimizer.tell(solutions)
if optimizer.should_stop():
breakThe full source code is available here.
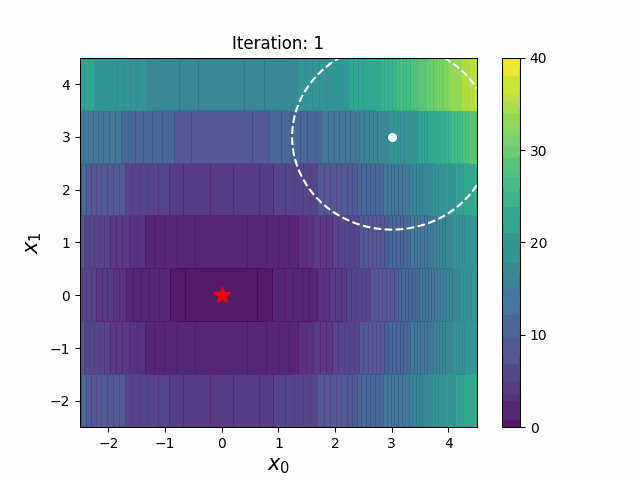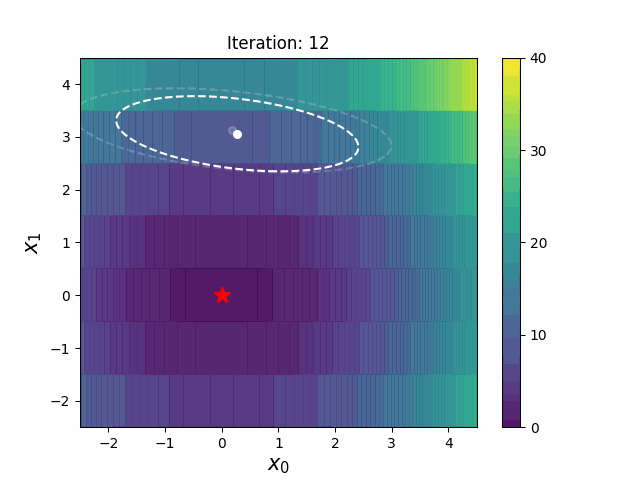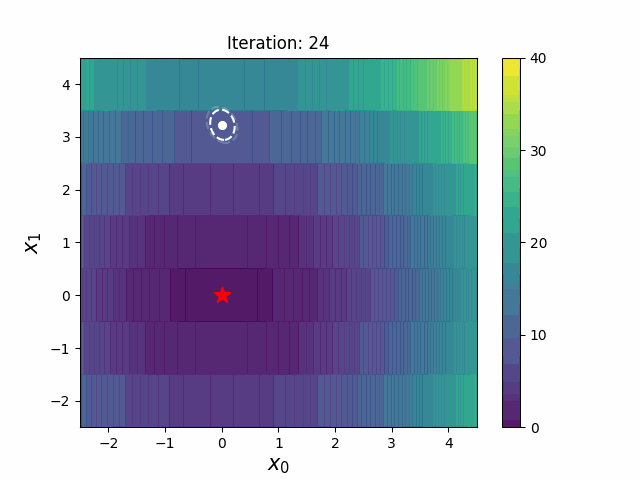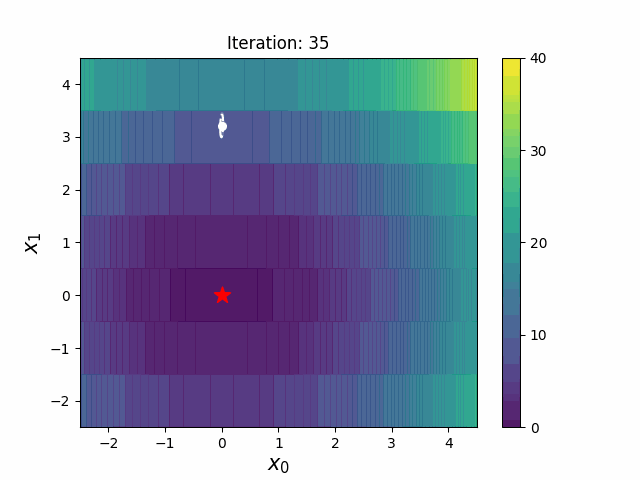
CMA-ES with Margin (CMAwM) introduces a lower bound on the marginal probability for each discrete dimension, ensuring that samples avoid being fixed to a single point. This method can be applied to mixed spaces consisting of continuous (such as float) and discrete elements (including integer and binary types).
| CMA | CMAwM |
|---|---|
 |
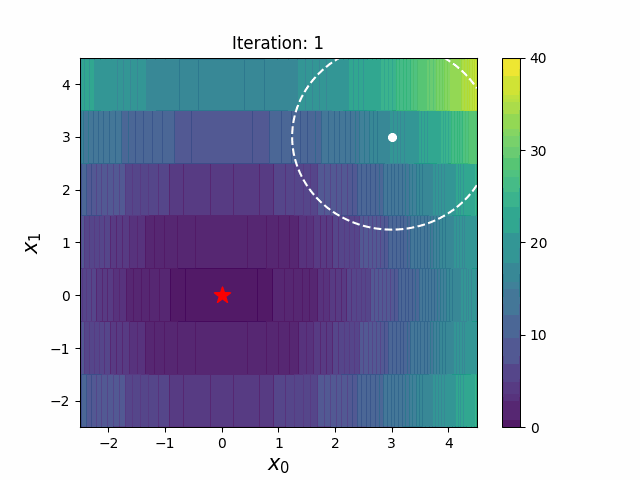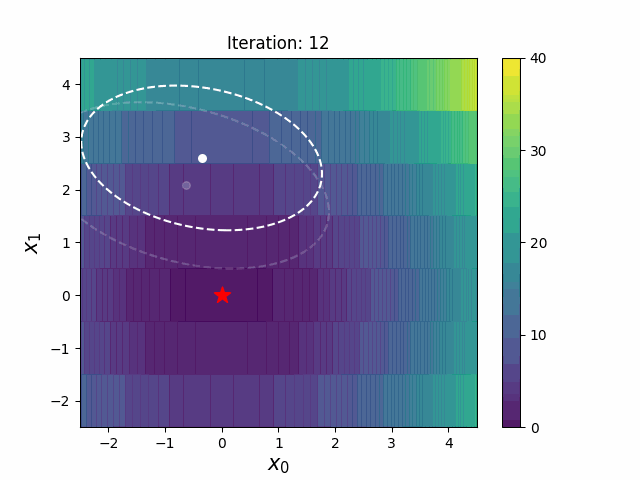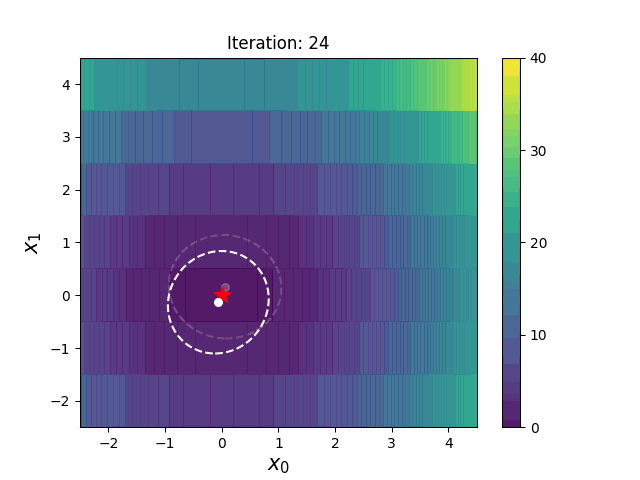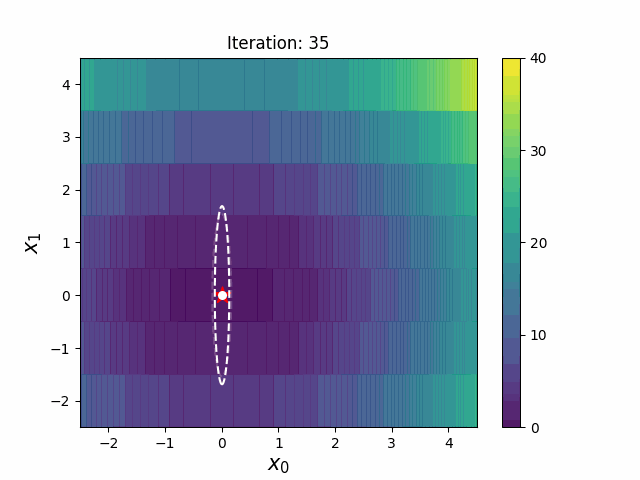 |
The above figures are taken from EvoConJP/CMA-ES_with_Margin.
Source code
import numpy as np
from cmaes import CMAwM
def ellipsoid_onemax(x, n_zdim):
n = len(x)
n_rdim = n - n_zdim
r = 10
if len(x) < 2:
raise ValueError("dimension must be greater one")
ellipsoid = sum([(1000 ** (i / (n_rdim - 1)) * x[i]) ** 2 for i in range(n_rdim)])
onemax = n_zdim - (0.0 < x[(n - n_zdim) :]).sum()
return ellipsoid + r * onemax
def main():
binary_dim, continuous_dim = 10, 10
dim = binary_dim + continuous_dim
bounds = np.concatenate(
[
np.tile([-np.inf, np.inf], (continuous_dim, 1)),
np.tile([0, 1], (binary_dim, 1)),
]
)
steps = np.concatenate([np.zeros(continuous_dim), np.ones(binary_dim)])
optimizer = CMAwM(mean=np.zeros(dim), sigma=2.0, bounds=bounds, steps=steps)
print(" evals f(x)")
print("====== ==========")
evals = 0
while True:
solutions = []
for _ in range(optimizer.population_size):
x_for_eval, x_for_tell = optimizer.ask()
value = ellipsoid_onemax(x_for_eval, binary_dim)
evals += 1
solutions.append((x_for_tell, value))
if evals % 300 == 0:
print(f"{evals:5d} {value:10.5f}")
optimizer.tell(solutions)
if optimizer.should_stop():
break
if __name__ == "__main__":
main()Source code is also available here.
Sep-CMA-ES is an algorithm that limits the covariance matrix to a diagonal form. This reduction in the number of parameters enhances scalability, making Sep-CMA-ES well-suited for high-dimensional optimization tasks. Additionally, the learning rate for the covariance matrix is increased, leading to superior performance over the (full-covariance) CMA-ES on separable functions.
Source code
import numpy as np
from cmaes import SepCMA
def ellipsoid(x):
n = len(x)
if len(x) < 2:
raise ValueError("dimension must be greater one")
return sum([(1000 ** (i / (n - 1)) * x[i]) ** 2 for i in range(n)])
if __name__ == "__main__":
dim = 40
optimizer = SepCMA(mean=3 * np.ones(dim), sigma=2.0)
print(" evals f(x)")
print("====== ==========")
evals = 0
while True:
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = ellipsoid(x)
evals += 1
solutions.append((x, value))
if evals % 3000 == 0:
print(f"{evals:5d} {value:10.5f}")
optimizer.tell(solutions)
if optimizer.should_stop():
breakFull source code is available here.
IPOP-CMA-ES is a method that involves restarting the CMA-ES with an incrementally increasing population size, as described below.
Source code
import math
import numpy as np
from cmaes import CMA
def ackley(x1, x2):
# https://www.sfu.ca/~ssurjano/ackley.html
return (
-20 * math.exp(-0.2 * math.sqrt(0.5 * (x1 ** 2 + x2 ** 2)))
- math.exp(0.5 * (math.cos(2 * math.pi * x1) + math.cos(2 * math.pi * x2)))
+ math.e + 20
)
if __name__ == "__main__":
bounds = np.array([[-32.768, 32.768], [-32.768, 32.768]])
lower_bounds, upper_bounds = bounds[:, 0], bounds[:, 1]
mean = lower_bounds + (np.random.rand(2) * (upper_bounds - lower_bounds))
sigma = 32.768 * 2 / 5 # 1/5 of the domain width
optimizer = CMA(mean=mean, sigma=sigma, bounds=bounds, seed=0)
for generation in range(200):
solutions = []
for _ in range(optimizer.population_size):
x = optimizer.ask()
value = ackley(x[0], x[1])
solutions.append((x, value))
print(f"#{generation} {value} (x1={x[0]}, x2 = {x[1]})")
optimizer.tell(solutions)
if optimizer.should_stop():
# popsize multiplied by 2 (or 3) before each restart.
popsize = optimizer.population_size * 2
mean = lower_bounds + (np.random.rand(2) * (upper_bounds - lower_bounds))
optimizer = CMA(mean=mean, sigma=sigma, population_size=popsize)
print(f"Restart CMA-ES with popsize={popsize}")Full source code is available here.
If you use our library in your work, please cite our paper:
Masahiro Nomura, Masashi Shibata.
cmaes : A Simple yet Practical Python Library for CMA-ES
https://arxiv.org/abs/2402.01373
Bibtex:
@article{nomura2024cmaes,
title={cmaes : A Simple yet Practical Python Library for CMA-ES},
author={Nomura, Masahiro and Shibata, Masashi},
journal={arXiv preprint arXiv:2402.01373},
year={2024}
}
For any questions, feel free to raise an issue or contact me at [email protected].
Projects using cmaes:
- Optuna : A hyperparameter optimization framework that supports CMA-ES using this library under the hood.
- Kubeflow/Katib : Kubernetes-based system for hyperparameter tuning and neural architecture search
- (If you are using
cmaesin your project and would like it to be listed here, please submit a GitHub issue.)
Other libraries:
We have great respect for all libraries involved in CMA-ES.
- pycma : Most renowned CMA-ES implementation, created and maintained by Nikolaus Hansen.
- pymoo : A library for multi-objective optimization in Python.
- evojax : evojax offers a JAX-port of this library.
- evosax : evosax provides a JAX-based implementation of CMA-ES and sep-CMA-ES, inspired by this library.
References:
- [Akiba et al. 2019] T. Akiba, S. Sano, T. Yanase, T. Ohta, M. Koyama, Optuna: A Next-generation Hyperparameter Optimization Framework, KDD, 2019.
- [Auger and Hansen 2005] A. Auger, N. Hansen, A Restart CMA Evolution Strategy with Increasing Population Size, CEC, 2005.
- [Hamano et al. 2022] R. Hamano, S. Saito, M. Nomura, S. Shirakawa, CMA-ES with Margin: Lower-Bounding Marginal Probability for Mixed-Integer Black-Box Optimization, GECCO, 2022.
- [Hansen 2016] N. Hansen, The CMA Evolution Strategy: A Tutorial. arXiv:1604.00772, 2016.
- [Nomura et al. 2021] M. Nomura, S. Watanabe, Y. Akimoto, Y. Ozaki, M. Onishi, Warm Starting CMA-ES for Hyperparameter Optimization, AAAI, 2021.
- [Nomura et al. 2023] M. Nomura, Y. Akimoto, I. Ono, CMA-ES with Learning Rate Adaptation: Can CMA-ES with Default Population Size Solve Multimodal and Noisy Problems?, GECCO, 2023.
- [Nomura and Shibata 2024] M. Nomura, M. Shibata, cmaes : A Simple yet Practical Python Library for CMA-ES, arXiv:2402.01373, 2024.
- [Ros and Hansen 2008] R. Ros, N. Hansen, A Simple Modification in CMA-ES Achieving Linear Time and Space Complexity, PPSN, 2008.
















