jgrss / geowombat Goto Github PK
View Code? Open in Web Editor NEWGeoWombat: Utilities for geospatial data
Home Page: https://geowombat.readthedocs.io
License: MIT License
GeoWombat: Utilities for geospatial data
Home Page: https://geowombat.readthedocs.io
License: MIT License
I just ran into an issue where polygon_to_array isn't replicating the x y coords of the xarray data example. Here's how I am resolving it, but maybe worth thinking about a better solution.
labels = gw.polygon_to_array(labels, col=col, data=data,band_name= [variable_name], fill=0)
np.all(data.x.values == category.x.values) & np.all(data.y.values == category.y.values)
False
labels = labels.assign_coords({"x": data.x.values, 'y':data.y.values})
np.all(data.x.values == labels.x.values) & np.all(data.y.values == labels.y.values)
True@rdenham if you have some spare time, test the new series class from the jax branch. I removed xarray and dask dependencies, so opening large images should be faster. Additionally, computation is on the GPU.
To test it:
Install geowombat from the jax branch.
Install jax with GPU support (command below is for CUDA 11.1)
pip install --upgrade "jax[cuda111]" -f https://storage.googleapis.com/jax-releases/jax_releases.html
Simple example to compute the mean over time:
import geowombat as gwCreate a custom class
class TemporalMean(gw.TimeModule):
def __init__(self):
super(TemporalMean, self).__init__()
def calculate(self, array):
"""Returns the window and the mean along the time axis"""
return array.mean(axis=0).squeeze()with gw.series(file_list, nodata=0, num_threads=4, window_size=(1024, 1024)) as src:
src.apply(TemporalMean(), outfile='mean.tif', bands=2, gain=0.0001, num_workers=4)I was just working on stuff for the book and ran into an odd issue on the most recent build of gw
import geowombat as gw
import geowombat as gw...
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
~/Documents/github/pyGIS/pygis/docs/test.py in
----> 645 import geowombat as gw
646 from geowombat.data import l8_224078_20200518
647
648 # with gw.config.update(sensor="bgr"):
649 with gw.open(l8_224078_20200518) as src:
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/geowombat/__init__.py in <module>
1 from . import config
----> 2 from .core.api import open
3 from .core import extract
4 from .core import sample
5 from .core import calc_area
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/geowombat/core/__init__.py in <module>
----> 1 from ..backends import transform_crs
2 from .io import apply
3 from .io import to_raster
4 from .io import to_netcdf
5 from .io import to_vrt
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/geowombat/backends/__init__.py in <module>
1 from .dask_ import Cluster
----> 2 from .xarray_ import concat, mosaic
3 from .xarray_ import warp_open
4 from .xarray_ import transform_crs
5
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/geowombat/backends/xarray_.py in <module>
5
6 from ..handler import add_handler
----> 7 from ..core.windows import get_window_offsets
8 from ..core.util import parse_filename_dates
9 from ..config import config
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/geowombat/core/windows.py in <module>
----> 1 from .util import n_rows_cols
2
3 from rasterio.windows import Window
4
5
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/geowombat/core/util.py in <module>
7
8 from ..handler import add_handler
----> 9 from ..moving import moving_window
10
11 import numpy as np
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/geowombat/moving/__init__.py in <module>
----> 1 from ._moving import moving_window
geowombat/moving/_moving.pyx in init geowombat.moving._moving()
ValueError: numpy.ndarray size changed, may indicate binary incompatibility. Expected 88 from C header, got 80 from PyObject
When you open an image using gw.config.update(refbounds=bound), any nodata values in the source data are converted to 0's.
Here is a reproducible example:
# create a dummy raster with nodata set to 255
from osgeo import gdal
from osgeo import osr
from rasterio.coords import BoundingBox
import numpy as np
ncol = 100
nrow = 100
egdata = np.random.randint(0,10, (nrow,ncol))
# add some nodata
egdata[0:10, 0:10] = 255
rasterOrigin = (1238095,-2305756)
pixelWidth = 10
pixelHeight = -10
originX = rasterOrigin[0]
originY = rasterOrigin[1]
driver = gdal.GetDriverByName('GTiff')
outRaster = driver.Create("test.tif", ncol, nrow, 1, gdal.GDT_Byte)
outRaster.SetGeoTransform((originX, pixelWidth, 0, originY, 0, pixelHeight))
outband = outRaster.GetRasterBand(1)
outband.WriteArray(egdata)
outband.SetNoDataValue(255)
outband.FlushCache()
outRasterSRS = osr.SpatialReference()
outRasterSRS.ImportFromEPSG(3577)
outRaster.SetProjection(outRasterSRS.ExportToWkt())
outband = None
outRaster = NoneNow, if you access this using gw:
with gw.config.update():
with gw.open("test.tif") as src:
data = src.data.compute()
print(data[0,0:10,0:10])you get the expected output, since these are all nodata values in the src
[[255 255 255 255 255 255 255 255 255 255]
[255 255 255 255 255 255 255 255 255 255]
[255 255 255 255 255 255 255 255 255 255]
[255 255 255 255 255 255 255 255 255 255]
[255 255 255 255 255 255 255 255 255 255]
[255 255 255 255 255 255 255 255 255 255]
[255 255 255 255 255 255 255 255 255 255]
[255 255 255 255 255 255 255 255 255 255]
[255 255 255 255 255 255 255 255 255 255]
[255 255 255 255 255 255 255 255 255 255]]
But if you use a bounds argument:
bounds = BoundingBox(left=rasterOrigin[0], bottom=rasterOrigin[1] - 60, right=rasterOrigin[0] + 60, top=rasterOrigin[1])
with gw.config.update(ref_bounds=bounds):
with gw.open("test.tif") as src:
data = src.data.compute()
print(data[0,0:10, 0:10])You get all 0's:
[[0 0 0 0 0 0]
[0 0 0 0 0 0]
[0 0 0 0 0 0]
[0 0 0 0 0 0]
[0 0 0 0 0 0]
[0 0 0 0 0 0]]
As a comparison, rasterio behaves as expected:
with rasterio.open("test.tif") as src:
rst = src.read(1, window=from_bounds(left=rasterOrigin[0], bottom=rasterOrigin[1] - 60, right=rasterOrigin[0] + 60, top=rasterOrigin[1], transform=src.transform))
print(rst[0:10, 0:10])[[255 255 255 255 255 255]
[255 255 255 255 255 255]
[255 255 255 255 255 255]
[255 255 255 255 255 255]
[255 255 255 255 255 255]
[255 255 255 255 255 255]]
I experimented with nodata in the config and open but didn't seem to make any difference.
In [68]: print(gw.__version__)
1.7.3
@mmann1123 the ML tests are failing for the same reason the docs are failing.
I opened an issue with sklearn-xarray folks about integration with geowombat data structures. They answered with a working example and I simultaneously figured most of it out and created a pull request to make it easier for us to use.
They want to create an example using gw data but worried most folks will have trouble generating the environment. I hate to ask you for something, but they are hoping for a dummy data set with some randomly created data to use as an example. I honestly have tried a few times and for many hours (and failed) to generate the same structure as:
``
with gw.open([rgbn_20160101, rgbn_20160401, rgbn_20160517],
band_names=['blue', 'green', 'red','nir'],
time_names=['t1', 't2','t3']) as src:
src
Any chance you know how to recreate that easily using xarray only? Might be useful for testing later anyways.
FYI @mmann1123, Dependabot broke the automatic release again. I'll try to work it out. In the meantime, if you have a PR that needs to bump any of major.minor.patch, it won't happen automatically.
I was just rebuilding docs for the book and noticed this. Just wondering if this needs to be handled.
with gw.config.update(scale_factor=0.0001):
with gw.open(l8_224078_20200518) as src:
print(src.attrs['scales'])
(1.0,1.0,1.0)
Morning!
First of all, thanks for the work, the library looks wicked!
I created a dedicated conda env by following your recommended steps:
pip install pip-tools
conda create -n geowombat python=3.7 cython scipy numpy zarr requests -c conda-forge
conda activate geowombat
sudo apt install libspatialindex-dev libgdal-dev
conda install -c conda-forge libspatialindex zarr requests
pip install git+https://github.com/jgrss/geowombat
python -c "import geowombat as gw;print(gw.__version__)"
Then I installed jupyterlab on top of the conda env to run the following:
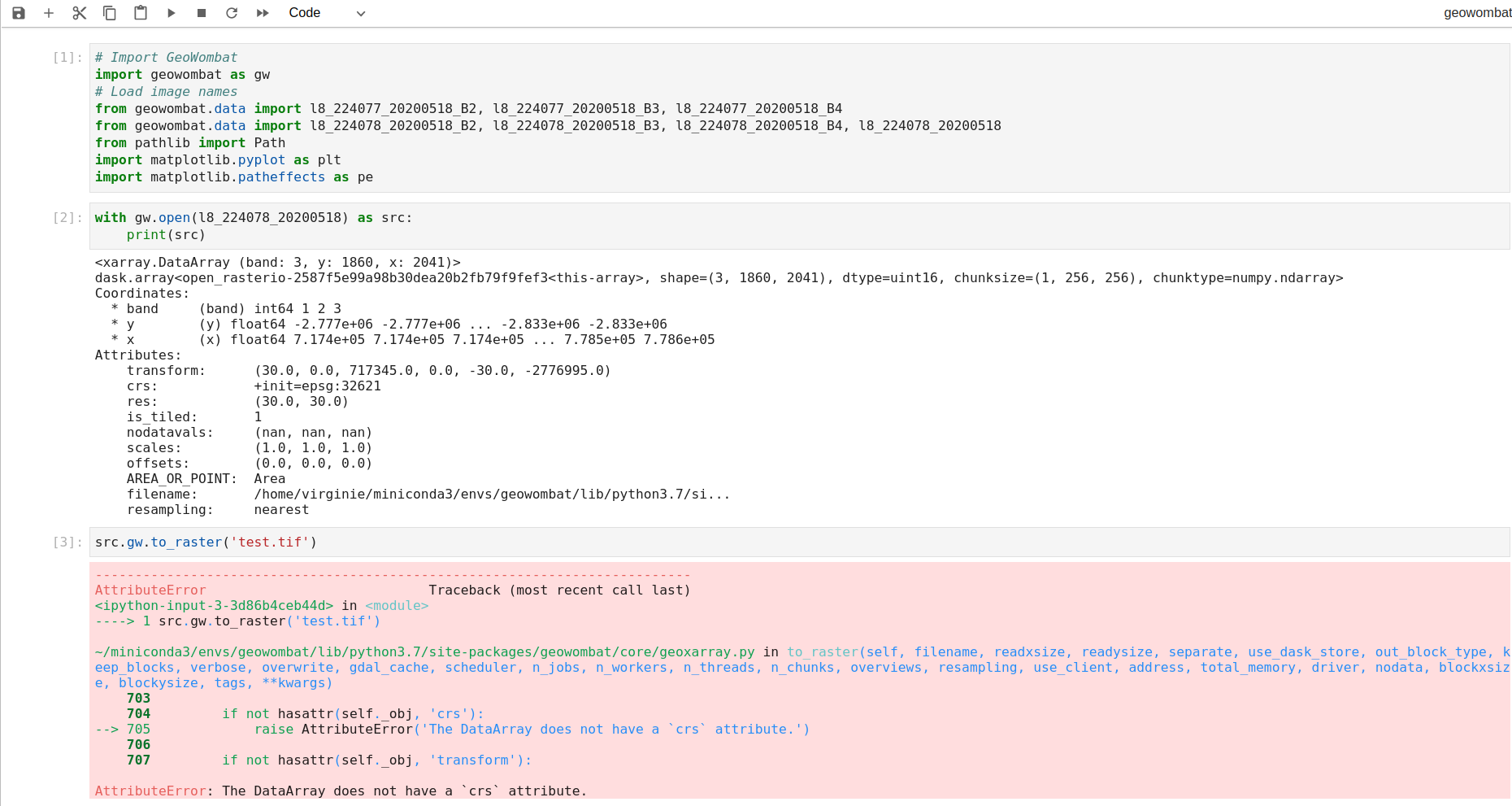
As you can see I am unable to save the raster despite I am using the example data available through the library. Same error message when trying to use some of my tif local files.
Any idea how to solve this bug? Thanks in advance!
Just got this report back from the conda feedstock or for details go here
Assuming pinning the xarray version pulls the older rasterio - breaking things downstream? What do you think @jgrss
The reported errors are:
- Encountered problems while solving:
- - package rasterio-1.0.23-py27h5b3f9e8_0 requires libgdal >=2.4.1,<2.5.0a0, but none of the providers can be installed
- - package pyproj-1.9.4-py27_0 requires python 2.7*, but none of the providers can be installed
Hi, I'm very excited about the prospects of geowombat making raster processing much easier! Thanks for doing this!
I'm still pretty new to Python, so maybe I am missing something with stacks. I'm trying to clip a raster stack but I get the 'open' object has no attribute 'crs' error. I tried using 'with' method as well as the code below and both return the same issue.
from geowombat.data import l8_224077_20200518_B2, l8_224077_20200518_B3,l8_224078_20200518_polygons
src= gw.open([l8_224077_20200518_B2, l8_224077_20200518_B3],
stack_dim="band",
band_names=["B2", "B3"])
srccl = gw.clip(src, l8_224078_20200518_polygons)
AttributeError Traceback (most recent call last)
File ~\Miniconda3\envs\gwenv\lib\site-packages\geowombat\core\sops.py:798, in SpatialOperations.clip(self, data, df, query, mask_data, expand_by)
796 try:
--> 798 if data.crs.strip() != CRS.from_dict(df_crs_).to_proj4().strip():
799 df = df.to_crs(data.crs)
AttributeError: 'open' object has no attribute 'crs'
During handling of the above exception, another exception occurred:
AttributeError Traceback (most recent call last)
Input In [62], in <cell line: 5>()
1 from geowombat.data import l8_224077_20200518_B2, l8_224077_20200518_B3,l8_224078_20200518_polygons
2 src= gw.open([l8_224077_20200518_B2, l8_224077_20200518_B3],
3 stack_dim="band",
4 band_names=["B2", "B3"])
----> 5 srccl = gw.clip(src, l8_224078_20200518_polygons)
File ~\Miniconda3\envs\gwenv\lib\site-packages\geowombat\core\sops.py:803, in SpatialOperations.clip(self, data, df, query, mask_data, expand_by)
799 df = df.to_crs(data.crs)
801 except:
--> 803 if data.crs.strip() != CRS.from_proj4(df_crs_).to_proj4().strip():
804 df = df.to_crs(data.crs)
806 row_chunks = data.gw.row_chunks
AttributeError: 'open' object has no attribute 'crs'
If I try the self method:
from geowombat.data import l8_224077_20200518_B2, l8_224077_20200518_B3,l8_224078_20200518_polygons
src= gw.open([l8_224077_20200518_B2, l8_224077_20200518_B3],
stack_dim="band",
band_names=["B2", "B3"])
srccl = src.gw.clip( l8_224078_20200518_polygons)
I get the error:
AttributeError Traceback (most recent call last)
Input In [66], in <cell line: 5>()
1 from geowombat.data import l8_224077_20200518_B2, l8_224077_20200518_B3,l8_224078_20200518_polygons
2 src= gw.open([l8_224077_20200518_B2, l8_224077_20200518_B3],
3 stack_dim="band",
4 band_names=["B2", "B3"])
----> 5 srccl = src.gw.clip( l8_224078_20200518_polygons)
AttributeError: 'open' object has no attribute 'gw'
Maybe I'm doing something incorrect with stacks, as I am not able to use to_raster with them either (added a note to #184 as well).
Thanks!
Paul
Xarray has removed support for the deprecated xarray.ufuncs functions since version v2022.06.rc0: https://docs.xarray.dev/en/stable/whats-new.html#breaking-changes.
Now I receive the following error:
import geowombat as gw
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Input In [2], in <cell line: 1>()
----> 1 import geowombat as gw
File /opt/tljh/user/envs/stable/lib/python3.8/site-packages/geowombat/__init__.py:5, in <module>
2 __version__ = '1.11.3'
4 from . import config
----> 5 from .core.api import open
6 from .core.api import load
7 from .core.api import series
File /opt/tljh/user/envs/stable/lib/python3.8/site-packages/geowombat/core/__init__.py:1, in <module>
----> 1 from ..backends import transform_crs
2 from .io import apply
3 from .io import to_raster
File /opt/tljh/user/envs/stable/lib/python3.8/site-packages/geowombat/backends/__init__.py:2, in <module>
1 from .dask_ import Cluster
----> 2 from .xarray_ import concat, mosaic
3 from .xarray_ import warp_open
4 from .xarray_ import transform_crs
File /opt/tljh/user/envs/stable/lib/python3.8/site-packages/geowombat/backends/xarray_.py:26, in <module>
24 import dask.array as da
25 import xarray as xr
---> 26 from xarray.ufuncs import maximum as xr_maximum
27 from xarray.ufuncs import minimum as xr_mininum
30 logger = logging.getLogger(__name__)
ModuleNotFoundError: No module named 'xarray.ufuncs'Searching the repo: https://github.com/jgrss/geowombat/search?q=xarray.ufuncs&type=code
Reveals at least one file needs updating:
geowombat/src/geowombat/backends/xarray_.py
Lines 26 to 27 in 342bb2b
Seems like the 'retry' package is now needed for gw 1.7.3, but it isn't listed in requirements.txt.
❯ python
Python 3.8.10 (default, Jun 2 2021, 10:49:15)
[GCC 9.4.0] on linux
Type "help", "copyright", "credits" or "license" for more information.
>>> import geowombat as gw
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File ".venv38gdal33/lib/python3.8/site-packages/geowombat/__init__.py", line 2, in <module>
from .core.api import open
File ".venv38gdal33/lib/python3.8/site-packages/geowombat/core/api.py", line 18, in <module>
from . import geoxarray
File ".venv38gdal33/lib/python3.8/site-packages/geowombat/core/geoxarray.py", line 19, in <module>
from ..util import imshow as gw_imshow
File ".venv38gdal33/lib/python3.8/site-packages/geowombat/util/__init__.py", line 2, in <module>
from .web import GeoDownloads
File ".venv38gdal33/lib/python3.8/site-packages/geowombat/util/web.py", line 14, in <module>
from ..radiometry import BRDF, LinearAdjustments, RadTransforms, landsat_pixel_angles, sentinel_pixel_angles, QAMasker, DOS, SixS
File ".venv38gdal33/lib/python3.8/site-packages/geowombat/radiometry/__init__.py", line 4, in <module>
from .sr import LinearAdjustments, RadTransforms, DOS
File ".venv38gdal33/lib/python3.8/site-packages/geowombat/radiometry/sr.py", line 11, in <module>
from .sixs import AOT, SixS
File ".venv38gdal33/lib/python3.8/site-packages/geowombat/radiometry/sixs.py", line 20, in <module>
from ..data import LUTDownloader, NASAEarthdataDownloader
File ".venv38gdal33/lib/python3.8/site-packages/geowombat/data/__init__.py", line 7, in <module>
from retry import retry
ModuleNotFoundError: No module named 'retry'
>>> exit()
Not a big issue, but is nice for installing if it is added as a requirement.
return the following error, even when using geowombat rgbn test image:
rasterio.errors.CRSError: Unable to open EPSG support file gcs.csv. Try setting the GDAL_DATA environment variable to point to the directory containing EPSG csv files.
GeoWombat now has a geowombat.norm_brdf function, but it hasn't been thoroughly tested.
Just wondering if you have any thoughts about this error with mpool. I tried making the chunks smaller, to no avail. After the stackerizer each chunk end up being (2152143, 42), which really isn't very large. Any tricks?
Also, would you prefer me ask these types of questions on git issue?
pt = ParallelTask(ds,
scheduler='mpool',
n_workers=2)
print('n chunks %s' % pt.n_chunks)
print('n windows %s' % pt.n_windows)
res = pt.map(user_func, 6 )
error Traceback (most recent call last)
~/Documents/github/ET_drought_2019/scripts/crop_loss.py in
146 print('n chunks %s' % pt.n_chunks)
147 print('n windows %s' % pt.n_windows)
---> 148 res = pt.map(user_func, 6 )
149
~/Documents/github/geowombat/geowombat/core/parallel.py in map(self, func, *args)
144 if self.scheduler == 'mpool':
145
--> 146 for result in tqdm(executor.imap_unordered(func, data_gen), total=n_windows_slice):
147 results.append(result)
148
~/anaconda3/envs/geowombatdebug/lib/python3.7/site-packages/tqdm/std.py in iter(self)
1163
1164 try:
-> 1165 for obj in iterable:
1166 yield obj
1167 # Update and possibly print the progressbar.
~/anaconda3/envs/geowombatdebug/lib/python3.7/multiprocessing/pool.py in next(self, timeout)
746 if success:
747 return value
--> 748 raise value
749
750 next = next # XXX
~/anaconda3/envs/geowombatdebug/lib/python3.7/multiprocessing/pool.py in _handle_tasks(taskqueue, put, outqueue, pool, cache)
429 break
430 try:
--> 431 put(task)
432 except Exception as e:
433 job, idx = task[:2]
~/anaconda3/envs/geowombatdebug/lib/python3.7/multiprocessing/connection.py in send(self, obj)
204 self._check_closed()
205 self._check_writable()
--> 206 self._send_bytes(_ForkingPickler.dumps(obj))
207
208 def recv_bytes(self, maxlength=None):
~/anaconda3/envs/geowombatdebug/lib/python3.7/multiprocessing/connection.py in _send_bytes(self, buf)
391 n = len(buf)
392 # For wire compatibility with 3.2 and lower
--> 393 header = struct.pack("!i", n)
394 if n > 16384:
395 # The payload is large so Nagle's algorithm won't be triggered
error: 'i' format requires -2147483648 <= number <= 2147483647
Trying to help resolve #216 but running into a likely easily fixed issue. As you suggested a while back I can do pip install --user -e ."[ml]" in my dockerfile. However I am getting an error now with the your sklearn fork. Maybe there's an easy work around?
RUN git clone https://github.com/jgrss/geowombat.git --branch jgrss/scales_216
RUN cd geowombat && pip install --user -e ."[ml]"
ERROR: Invalid requirement: 'sklearn-xarray@ git+https://github.com/jgrss/sklearn-xarray.git; extra == "ml"'
The command '/bin/sh -c cd geowombat && pip install --user -e ."[ml]"' returned a non-zero code: 1
I started a new conda env and once again have GDAL path issues. I noticed that fiona seems to have resolved this internally, and thinking maybe we could implement their solution, see link below. Wish I could be of help on this, but this is way above my pay grade.
CPLE_OpenFailedError: Unable to open EPSG support file gcs.csv. Try setting the GDAL_DATA environment variable to point to the directory containing EPSG csv files.
Hello, I tried writing data arrays using the to_raster function. But, I am getting an error 'TypeError: tqdm.std.tqdm() argument after ** must be a mapping, not NoneType'. I don't know what I am doing wrong so I appreciate if you can look into it.
Ok after a lot of fumbling around, I think I figured out an issue I have been having. It seems like in data, window_id, num_workers = list(itertools.chain(*args)) , the window_id is actually the chunk_id. I came across this because of the unusual size of my data, but managed to finally figure out a way to replicate it.
If n_windows is ever larger than n_chunks, the window_id iterates through values of n_chunks, but then repeats itself until you reach window_id. So in my case I keep overwriting my file.
Here's an example
import numpy as np
import dask.array as da
import xarray as xr
from geowombat.core.parallel import ParallelTask
import geowombat as gw
import itertools
import os
sys.path.append('/home/mmann1123/Documents/github/xr_fresh/')
from xr_fresh.utils import save_pickle,open_pickle, unique,add_categorical,add_time_targets,compressed_pickle,decompress_pickle
from xr_fresh.transformers import Stackerizer
from geowombat.data import rgbn_20160101, rgbn_20160401, rgbn_20160517
out_folder = '~/Desktop/'
def user_func(*args):
data, window_id, num_workers = list(itertools.chain(*args)) # num_workers=1
X = Stackerizer(stack_dims = ('y','x','time'), direction='stack').fit_transform(data) # NOTE stack y before x!!!
X = X.to_pandas()
X.columns = X.columns.astype(str)
X.to_parquet(out_folder+'/testing/test_'+ '_%05d.parquet' % window_id)
return None
multiplier = 50
import geowombat as gw
with gw.open([rgbn_20160101, rgbn_20160401, rgbn_20160517]*multiplier,
band_names=['blue', 'green', 'red','nir'],
time_names=[str(x) for x in range(multiplier*3)],chunks=25) as src:
print(src)
print('stack:')
pt = ParallelTask(src,
scheduler='threads',
n_workers=3)
print('n chunks %s' % pt.n_chunks)
print('n windows %s' % pt.n_windows)
res = pt.map(user_func, 4) Ran into an issue when mosaicing a single band image and writing to vrt. Sorry not quite sure how to address it. If you point me in the right direction I can try to figure it out. For a working example see a nearly identical dataset here [here](https://drive.google.com/file/d/1xHPASY0ZT40rTvS8Uc866FJvpKwbWn1S/view?usp=sharing
https://drive.google.com/file/d/1uoJI7Hl1HFdW6MD6paBR_afWwh6jaZsG/view?usp=sharing
https://drive.google.com/file/d/1HYPauHCLyj5kVQMDbQtZa-BvEgHmIIDl/view?usp=sharing
https://drive.google.com/file/d/1lb_-yUoYpu3CCbl3nfAmPW_sT6jj3Zr-/view?usp=sharing)
`with gw.open(glob('./data/NDWI/2020tif'), band_names=['NDWI'],
time_names=['01-01-2020'],
bounds_by='union',mosaic=True,
num_workers=5) as ds:
print(ds)
#ds.sel(band=['NDWI']).gw.imshow(robust=True)
# Write the data to a VRT
gw.to_vrt(data=ds, filename='./data/NDWI/NDWI_01-01-2020.vrt', nodata=0.0)
File "", line 12, in
gw.to_vrt(data=ds, filename='./data/NDWI/NDWI_01-01-2020.vrt', nodata=0.0)
File "/home/mmann1123/anaconda3/envs/geowombat/lib/python3.7/site-packages/geowombat/core/io.py", line 366, in to_vrt
with rio.open(data.filename) as src:
File "/home/mmann1123/anaconda3/envs/geowombat/lib/python3.7/site-packages/xarray/core/common.py", line 233, in getattr
"{!r} object has no attribute {!r}".format(type(self).name, name)
AttributeError: 'DataArray' object has no attribute 'filename'
`
When trying to extract data using a tuple of bounded coordinates, I get an error. Here is the example taken from the documentation:
bounds = (793000.0, 2049000.0, 794000.0, 2050000.0)
with gw.open(rgbn,
band_names=['green', 'red', 'nir'],
num_workers=8,
indexes=[2, 3, 4],
bounds=bounds,
out_dtype='float32') as ds:
print(ds)
And the error:
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
~/anaconda3/envs/gwenv/lib/python3.7/site-packages/numpy/core/function_base.py in linspace(start, stop, num, endpoint, retstep, dtype, axis)
116 try:
--> 117 num = operator.index(num)
118 except TypeError:
TypeError: 'float' object cannot be interpreted as an integer
During handling of the above exception, another exception occurred:
TypeError Traceback (most recent call last)
<ipython-input-18-26e292b10c6b> in <module>
6 indexes=[2, 3, 4],
7 bounds=bounds,
----> 8 out_dtype='float32') as ds:
9 print(ds)
~/anaconda3/envs/gwenv/lib/python3.7/contextlib.py in __enter__(self)
110 del self.args, self.kwds, self.func
111 try:
--> 112 return next(self.gen)
113 except StopIteration:
114 raise RuntimeError("generator didn't yield") from None
~/anaconda3/envs/gwenv/lib/python3.7/site-packages/geowombat/core/api.py in open(filename, return_as, band_names, time_names, stack_dim, bounds, bounds_by, resampling, mosaic, overlap, dtype, num_workers, **kwargs)
348 bounds=bounds,
349 num_workers=num_workers,
--> 350 **kwargs)
351
352 else:
~/anaconda3/envs/gwenv/lib/python3.7/site-packages/geowombat/core/api.py in read(filename, band_names, time_names, bounds, num_workers, **kwargs)
162 kwargs['window'] = from_bounds(*bounds, transform=src.transform)
163
--> 164 ycoords, xcoords, attrs = get_attrs(src, **kwargs)
165
166 data = dask.compute(read_delayed(filename, 0, **kwargs),
~/anaconda3/envs/gwenv/lib/python3.7/site-packages/geowombat/core/api.py in get_attrs(src, **kwargs)
35 ycoords = np.linspace(src.bounds.top - (kwargs['window'].row_off * src.res[0]),
36 src.bounds.top - (kwargs['window'].row_off * src.res[0]) -
---> 37 (kwargs['window'].height * src.res[0]), kwargs['window'].height)
38
39 xcoords = np.linspace(src.bounds.left + (kwargs['window'].col_off * src.res[0]),
<__array_function__ internals> in linspace(*args, **kwargs)
~/anaconda3/envs/gwenv/lib/python3.7/site-packages/numpy/core/function_base.py in linspace(start, stop, num, endpoint, retstep, dtype, axis)
119 raise TypeError(
120 "object of type {} cannot be safely interpreted as an integer."
--> 121 .format(type(num)))
122
123 if num < 0:
TypeError: object of type <class 'float'> cannot be safely interpreted as an integer.
This is caused in the call to numpy.linspace using the calculated window width as the argument num. But the window width as calculated by rio.window.from_bounds returns a float not an integer.
Perhaps this should be cast to an integer in the linspace call?
My pip freeze output:
affine==2.3.0
alabaster==0.7.12
asciitree==0.3.3
attrs==19.3.0
Babel==2.8.0
backcall==0.1.0
bleach==3.1.0
certifi==2019.11.28
cftime==1.0.4.2
chardet==3.0.4
Click==7.0
click-plugins==1.1.1
cligj==0.5.0
cloudpickle==1.2.2
cycler==0.10.0
Cython==0.29.14
dask==2.9.2
dask-glm==0.2.0
dask-ml==1.2.0
dateparser==0.7.2
decorator==4.4.1
defusedxml==0.6.0
Deprecated==1.2.7
distributed==2.9.3
docutils==0.16
entrypoints==0.3
fasteners==0.15
Fiona==1.8.13
fsspec==0.6.2
GDAL==2.4.0
geopandas==0.6.2
geowombat==1.0.7
h5netcdf==0.7.4
h5py==2.10.0
HeapDict==1.0.1
idna==2.8
imagesize==1.2.0
importlib-metadata==1.4.0
ipykernel==5.1.3
ipython==7.11.1
ipython-genutils==0.2.0
jedi==0.16.0
Jinja2==2.10.3
joblib==0.14.1
json5==0.8.5
jsonschema==3.2.0
jupyter-client==5.3.4
jupyter-core==4.6.1
jupyterlab==1.2.6
jupyterlab-server==1.0.6
kiwisolver==1.1.0
latexcodec==2.0.0
llvmlite==0.31.0
locket==0.2.0
MarkupSafe==1.1.1
matplotlib==3.1.2
mistune==0.8.4
mkl-fft==1.0.15
mkl-random==1.1.0
mkl-service==2.3.0
monotonic==1.5
msgpack==0.6.2
multipledispatch==0.6.0
munch==2.5.0
nbconvert==5.6.1
nbformat==5.0.4
netCDF4==1.5.3
notebook==6.0.3
numba==0.47.0
numcodecs==0.6.4
numexpr==2.7.1
numpy==1.18.1
numpydoc==0.9.2
opencv-python==4.1.2.30
oset==0.1.3
packaging==20.1
pandas==0.25.3
pandocfilters==1.4.2
parso==0.5.2
partd==1.1.0
pexpect==4.8.0
pickleshare==0.7.5
prometheus-client==0.7.1
prompt-toolkit==3.0.2
psutil==5.6.7
ptyprocess==0.6.0
pybtex==0.22.2
pybtex-docutils==0.2.2
Pygments==2.5.2
pyparsing==2.4.6
pyproj==2.4.2.post1
pyrsistent==0.15.7
python-dateutil==2.8.1
pytz==2019.3
PyYAML==5.3
pyzmq==18.1.1
rasterio==1.1.2
regex==2020.1.8
requests==2.22.0
scikit-learn==0.22.1
scipy==1.4.1
Send2Trash==1.5.0
Shapely==1.6.4.post2
six==1.14.0
snowballstemmer==2.0.0
snuggs==1.4.7
sortedcontainers==2.1.0
Sphinx==2.3.1
sphinx-automodapi==0.12
sphinxcontrib-applehelp==1.0.1
sphinxcontrib-bibtex==1.0.0
sphinxcontrib-devhelp==1.0.1
sphinxcontrib-htmlhelp==1.0.2
sphinxcontrib-jsmath==1.0.1
sphinxcontrib-qthelp==1.0.2
sphinxcontrib-serializinghtml==1.1.3
tblib==1.6.0
terminado==0.8.3
testpath==0.4.4
toolz==0.10.0
tornado==6.0.3
tqdm==4.42.0
traitlets==4.3.3
tzlocal==2.0.0
urllib3==1.25.8
wcwidth==0.1.8
webencodings==0.5.1
wget==3.2
wrapt==1.11.2
xarray==0.14.1
zarr==2.4.0
zict==1.0.0
zipp==2.1.0affine==2.3.0
alabaster==0.7.12
asciitree==0.3.3
attrs==19.3.0
Babel==2.8.0
backcall==0.1.0
bleach==3.1.0
certifi==2019.11.28
cftime==1.0.4.2
chardet==3.0.4
Click==7.0
click-plugins==1.1.1
cligj==0.5.0
cloudpickle==1.2.2
cycler==0.10.0
Cython==0.29.14
dask==2.9.2
dask-glm==0.2.0
dask-ml==1.2.0
dateparser==0.7.2
decorator==4.4.1
defusedxml==0.6.0
Deprecated==1.2.7
distributed==2.9.3
docutils==0.16
entrypoints==0.3
fasteners==0.15
Fiona==1.8.13
fsspec==0.6.2
GDAL==2.4.0
geopandas==0.6.2
geowombat==1.0.7
h5netcdf==0.7.4
h5py==2.10.0
HeapDict==1.0.1
idna==2.8
imagesize==1.2.0
importlib-metadata==1.4.0
ipykernel==5.1.3
ipython==7.11.1
ipython-genutils==0.2.0
jedi==0.16.0
Jinja2==2.10.3
joblib==0.14.1
json5==0.8.5
jsonschema==3.2.0
jupyter-client==5.3.4
jupyter-core==4.6.1
jupyterlab==1.2.6
jupyterlab-server==1.0.6
kiwisolver==1.1.0
latexcodec==2.0.0
llvmlite==0.31.0
locket==0.2.0
MarkupSafe==1.1.1
matplotlib==3.1.2
mistune==0.8.4
mkl-fft==1.0.15
mkl-random==1.1.0
mkl-service==2.3.0
monotonic==1.5
msgpack==0.6.2
multipledispatch==0.6.0
munch==2.5.0
nbconvert==5.6.1
nbformat==5.0.4
netCDF4==1.5.3
notebook==6.0.3
numba==0.47.0
numcodecs==0.6.4
numexpr==2.7.1
numpy==1.18.1
numpydoc==0.9.2
opencv-python==4.1.2.30
oset==0.1.3
packaging==20.1
pandas==0.25.3
pandocfilters==1.4.2
parso==0.5.2
partd==1.1.0
pexpect==4.8.0
pickleshare==0.7.5
prometheus-client==0.7.1
prompt-toolkit==3.0.2
psutil==5.6.7
ptyprocess==0.6.0
pybtex==0.22.2
pybtex-docutils==0.2.2
Pygments==2.5.2
pyparsing==2.4.6
pyproj==2.4.2.post1
pyrsistent==0.15.7
python-dateutil==2.8.1
pytz==2019.3
PyYAML==5.3
pyzmq==18.1.1
rasterio==1.1.2
regex==2020.1.8
requests==2.22.0
scikit-learn==0.22.1
scipy==1.4.1
Send2Trash==1.5.0
Shapely==1.6.4.post2
six==1.14.0
snowballstemmer==2.0.0
snuggs==1.4.7
sortedcontainers==2.1.0
Sphinx==2.3.1
sphinx-automodapi==0.12
sphinxcontrib-applehelp==1.0.1
sphinxcontrib-bibtex==1.0.0
sphinxcontrib-devhelp==1.0.1
sphinxcontrib-htmlhelp==1.0.2
sphinxcontrib-jsmath==1.0.1
sphinxcontrib-qthelp==1.0.2
sphinxcontrib-serializinghtml==1.1.3
tblib==1.6.0
terminado==0.8.3
testpath==0.4.4
toolz==0.10.0
tornado==6.0.3
tqdm==4.42.0
traitlets==4.3.3
tzlocal==2.0.0
urllib3==1.25.8
wcwidth==0.1.8
webencodings==0.5.1
wget==3.2
wrapt==1.11.2
xarray==0.14.1
zarr==2.4.0
zict==1.0.0
zipp==2.1.0
Currently, geowombat.extract iterates over the output of dask.vindex using Python loops. This behavior should be changed to use numpy reshaping.
Hi dear
When trying to open my tiff file, I get this error: ValueError: negative dimensions are not allowed
code:
with gw.open(mytif) as src:
print(src)
File "/home/---/.local/lib/python3.8/site-packages/geowombat/core/api.py", line 493, in init
self.data = warp_open(
File "/home/---/.local/lib/python3.8/site-packages/geowombat/backends/xarray_.py", line 271, in warp_open
with open_rasterio(
File "/home/---/.local/lib/python3.8/site-packages/geowombat/backends/xarray_rasterio_.py", line 316, in open_rasterio
x, _ = riods.transform * (np.arange(nx) + 0.5, np.zeros(nx) + 0.5)
ValueError: negative dimensions are not allowed
The same tiff file can be read easily using rasterio, gdal... by with geowombat it's not possible
Thanks
I just ran into a case where generating the x and y coords doesn't match the original xarray dimensions. I can share the file if you want to see it.
When running polygon_to_array:
ds.shape
6681 by 5172
after generating coordinates:
xcoords = np.arange(left + cellxh, left + cellxh + dst_width * abs(cellx), cellx)
ycoords = np.arange(top - cellyh, top - cellyh - dst_height * abs(celly), -celly)
len(xcoords)
6682
gdalinfo --version
GDAL 3.3.2, released 2021/09/01
pyenv virtualenv 3.8.12 venv.gw
pyenv activate venv.gw
(venv.gw) pip install GDAL==3.3.2
(venv.gw) python -c "from osgeo import gdal;print(gdal.__version__)"
3.3.2(venv.gw) pip install git+https://github.com/jgrss/geowombat.git
(venv.gw) pip install git+https://github.com/jgrss/geowombat.git#egg=project[stac]
(venv.gw) pip install "geowombat[stac]@git+https://github.com/jgrss/geowombat.git"
(venv.gw) pip install "geowombat[ml,coreg,stac,perf,tests]@git+https://github.com/jgrss/geowombat.git"
For some reason, the command above initiates a Python GDAL build even when Python GDAL is already installed. For example, an attempted build for GDAL==3.5.* occurs even though I have GDAL=3.3.2 installed.
(venv.gw) pip install "geowombat[ml,stac,perf,tests]@git+https://github.com/jgrss/geowombat.git"
This issue stems from arosics, which has a gdal requirement. However, there is no GDAL version requirement specified, so I am not sure why pip is attempting to install a higher version.
First, arosics will be removed from the 'coreg' extra list in geowombat. Then, using the following command, all dependencies can be installed without pip installing GDAL.
(venv.gw) pip install arosics>=1.7.8 --no-deps && pip install "geowombat[coreg]@git+https://github.com/jgrss/geowombat.git"
pip install "geowombat[extra]@git+https://github.com/jgrss/geowombat.git" extra install format.setup.cfg to remove arosics.pip install arosics>=1.7.8 --no-deps && pip install "geowombat[coreg]@git+https://github.com/jgrss/geowombat.git".I am trying to debug this, but having some trouble. Although working previously polygon_to_array is having trouble. I tried a few polygons and I am getting the same issue. Just wondering if you have the same issue.
with gw.open(tiffs, time_names = time_names) as ds:
print(ds)
<Client: 'inproc://192.168.86.77/236471/8' processes=1 threads=12, memory=67.08 GB>
go to http://localhost:8787/status for dask dashboard
<xarray.DataArray (time: 7, band: 38, y: 2586, x: 3339)>
dask.array<concatenate, shape=(7, 38, 2586, 3339), dtype=float64, chunksize=(1, 1, 512, 512), chunktype=numpy.ndarray>
Coordinates:
* band (band) int64 1 2 3 4 5 6 7 8 9 10 ... 29 30 31 32 33 34 35 36 37 38
* y (y) float64 9.209 9.207 9.204 9.202 9.2 ... 3.41 3.408 3.406 3.403
* x (x) float64 40.5 40.5 40.51 40.51 40.51 ... 47.99 47.99 48.0 48.0
* time (time) <U4 '2010' '2011' '2012' '2013' '2014' '2015' '2016'
Attributes:
transform: (0.002245788210298804, 0.0, 40.500544584528626, 0.0, -0.0...
crs: +init=epsg:4326
res: (0.002245788210298804, 0.002245788210298804)
is_tiled: 1
nodatavals: (0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
scales: (1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1...
offsets: (0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0...
filename: ['y000002586_x000003341_h000002586_w000003340.tif', 'y000...
resampling: nearest
AREA_OR_POINT: Area labels = gw.polygon_to_array(labels, data=ds )
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
~/Documents/github/ET_drought_2019/scripts/crop_loss.py in
90 labels['band'] = [variable_name]
91 else:
---> 92 labels = gw.polygon_to_array(labels, col=col, data=data )
93 labels['band'] = [variable_name]
94
~/Documents/github/geowombat/geowombat/core/conversion.py in polygon_to_array(self, polygon, col, data, cellx, celly, band_name, row_chunks, col_chunks, src_res, fill, default_value, all_touched, dtype, sindex)
576
577 # Get intersecting features
--> 578 int_idx = sorted(list(sindex.intersection(tuple(data.gw.geodataframe.total_bounds.flatten()))))
579
580 if not int_idx:
~/Documents/github/geowombat/geowombat/core/properties.py in geodataframe(self)
811 return gpd.GeoDataFrame(data=[1],
812 columns=['grid'],
--> 813 geometry=[self.geometry],
814 crs=self._obj.crs)
815
~/Documents/github/geowombat/geowombat/core/properties.py in geometry(self)
766 """
767
--> 768 return Polygon([(self.left, self.bottom),
769 (self.left, self.top),
770 (self.right, self.top),
~/Documents/github/geowombat/geowombat/core/properties.py in left(self)
659 """
660
--> 661 return float(self._obj.x.min().values) - self.cellxh
662
663 @property
AttributeError: 'NoneType' object has no attribute 'x'Google Cloud public reference (Landsat)
Google Cloud public reference (Sentinel-2)
Landsat 8 Collection 1
Read file from Google example
@jgrss For the machine learning example in the tutorial I was getting the following:
import geowombat as gw
from geowombat.data import l8_224078_20200518, l8_224078_20200518_polygons
from geowombat.ml import fit
import geopandas as gpd
from sklearn_xarray.preprocessing import Featurizer
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import LabelEncoder, StandardScaler
from sklearn.decomposition import PCA
from sklearn.naive_bayes import GaussianNB
le = LabelEncoder()
# The labels are string names, so here we convert them to integers
labels = gpd.read_file(l8_224078_20200518_polygons)
labels['lc'] = le.fit(labels.name).transform(labels.name)
# Use a data pipeline
pl = Pipeline([('featurizer', Featurizer()),
('scaler', StandardScaler()),
('pca', PCA()),
('clf', GaussianNB())])
# Fit the classifier
with gw.open(l8_224078_20200518, chunks=128) as src:
X, clf = fit(data= src, labels=labels, clf=pl, col='lc')
print(clf)
print(X)
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
~/Documents/github/pyGIS/pygis/docs/test.py in <module>
1874 # Fit the classifier
1875 with gw.open(l8_224078_20200518, chunks=128) as src:
---> 1876 X, clf = fit(data= src, labels=labels, clf=pl, col='lc')
1877
1878 print(clf)
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/geowombat/ml/classifiers.py in fit(self, data, labels, clf, grid_search, targ_name, targ_dim_name, col)
199 """
200
--> 201 data = self._prepare_labels(data, labels, col, targ_name)
202 X, Xna = self._prepare_predictors(data, targ_name)
203 clf = self._prepare_classifiers(clf)
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/geowombat/ml/classifiers.py in _prepare_labels(data, labels, col, targ_name)
67 labels = labels.where(labels != 0)
68
---> 69 data.coords[targ_name] = (['time', 'y', 'x'], labels)
70
71 return data
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/xarray/core/coordinates.py in __setitem__(self, key, value)
39
40 def __setitem__(self, key: Hashable, value: Any) -> None:
---> 41 self.update({key: value})
42
43 @property
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/xarray/core/coordinates.py in update(self, other)
162 other_vars = getattr(other, "variables", other)
163 coords, indexes = merge_coords(
--> 164 [self.variables, other_vars], priority_arg=1, indexes=self.xindexes
165 )
166 self._update_coords(coords, indexes)
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/xarray/core/merge.py in merge_coords(objects, compat, join, priority_arg, indexes, fill_value)
470 coerced, join=join, copy=False, indexes=indexes, fill_value=fill_value
471 )
--> 472 collected = collect_variables_and_indexes(aligned)
473 prioritized = _get_priority_vars_and_indexes(aligned, priority_arg, compat=compat)
474 variables, out_indexes = merge_collected(collected, prioritized, compat=compat)
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/xarray/core/merge.py in collect_variables_and_indexes(list_of_mappings)
292 append_all(coords, indexes)
293
--> 294 variable = as_variable(variable, name=name)
295 if variable.dims == (name,):
296 variable = variable.to_index_variable()
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/xarray/core/variable.py in as_variable(obj, name)
120 if isinstance(obj[1], DataArray):
121 raise TypeError(
--> 122 "Using a DataArray object to construct a variable is"
123 " ambiguous, please extract the data using the .data property."
124 )
TypeError: Using a DataArray object to construct a variable is ambiguous, please extract the data using the .data property.I am again seeing some oddities with extract. Wondering if you have any ideas for problem solving it, just not sure where to start. I have a large vector dataset of 70k administrative units in Ethiopia, trying to get monthly precip means for each unit. Precip is from CHIRPs and is 0.05 dd res.
Example image, resampled to 0.01dd.

Open and extract code:
with gw.config.update(ref_bounds=bounds, ref_res=(0.01, 0.01)):
with gw.open(
f_list,
band_names=["ppt"],
time_names=dates,
nodata=-9999,
resampling="bilinear",
) as ds:
print(ds)
df = ds.gw.extract(
aoi=eas,
all_touched=True,
band_names=ds.band.values.tolist(),
time_names=ds.time.values.tolist(),
n_threads=4, # n_jobs creates memory error that is uncaught
verbose=2,
)
print(df.head)
ag = df.groupby(by=["id"]) # ive also tried aggregating by the administrative code, no change
.agg("mean")
)
1st issue, without resampling to a higher resolution say 0.01 dd, 30k+ administrative units return no data, even with 'all touched' as true.
2nd issue, if I do resample, my outputted admin units take on a really weird block pattern. I have also tried the groupby based on the unique code for the admin unit, although id seems to correspond to the index on the features. This is the same for resample = bilinear and nearest.
Hey Jordan, I am trying to extract values to a large set of polygons. Just not sure how to interpret. Any ideas? I am using the lastest git version.
with gw.config.update(ref_image=example, ref_bounds=bounds):
with gw.open(
files,
band_names=["ppt_mean", "ppt_sum"],
stack_dim="band",
nodata=np.nan,
) as ds:
df = ds.gw.extract(
EAs,
all_touched=True,
band_names=["ppt_mean", "ppt_sum"],
time_names=[date],
n_jobs=1,
)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
~/Documents/github/ET_Random_James_Tasks/ET_precip_2012_2019.py in <module>
4 # band_names=["ppt_mean", "ppt_sum"],
5 # time_names=[date],
----> 6 n_jobs=1,
7 )
~/anaconda3/envs/geowombat2/lib/python3.7/site-packages/geowombat/core/geoxarray.py in extract(self, aoi, bands, time_names, band_names, frac, all_touched, mask, n_jobs, verbose, **kwargs)
1217 n_jobs=n_jobs,
1218 verbose=verbose,
-> 1219 **kwargs)
1220
1221 def band_mask(self, valid_bands, src_nodata=None, dst_clear_val=0, dst_mask_val=1):
~/anaconda3/envs/geowombat2/lib/python3.7/site-packages/geowombat/core/sops.py in extract(self, data, aoi, bands, time_names, band_names, frac, min_frac_area, all_touched, id_column, time_format, mask, n_jobs, verbose, n_workers, n_threads, use_client, address, total_memory, processes, pool_kwargs, **kwargs)
655 n_jobs=n_jobs,
656 verbose=verbose,
--> 657 **pool_kwargs)
658
659 if df.empty:
~/anaconda3/envs/geowombat2/lib/python3.7/site-packages/geowombat/core/conversion.py in prepare_points(self, data, aoi, frac, min_frac_area, all_touched, id_column, mask, n_jobs, verbose, **kwargs)
418 id_column=id_column,
419 n_jobs=n_jobs,
--> 420 **kwargs)
421
422 if not df.empty:
~/anaconda3/envs/geowombat2/lib/python3.7/site-packages/geowombat/core/conversion.py in polygons_to_points(data, df, frac, min_frac_area, all_touched, id_column, n_jobs, **kwargs)
476 meta,
477 frac,
--> 478 min_frac_area)
479
480 if not point_df.empty:
~/anaconda3/envs/geowombat2/lib/python3.7/site-packages/geowombat/core/util.py in sample_feature(df_row, id_column, df_columns, crs, res, all_touched, meta, frac, min_frac_area, feature_array)
553
554 for col in other_cols:
--> 555 fea_df.loc[:, col] = df_row[col]
556
557 return fea_df
~/anaconda3/envs/geowombat2/lib/python3.7/site-packages/pandas/core/indexing.py in __setitem__(self, key, value)
690
691 iloc = self if self.name == "iloc" else self.obj.iloc
--> 692 iloc._setitem_with_indexer(indexer, value, self.name)
693
694 def _validate_key(self, key, axis: int):
~/anaconda3/envs/geowombat2/lib/python3.7/site-packages/pandas/core/indexing.py in _setitem_with_indexer(self, indexer, value, name)
1586 if not is_list_like_indexer(value):
1587 raise ValueError(
-> 1588 "cannot set a frame with no "
1589 "defined index and a scalar"
1590 )
ValueError: cannot set a frame with no defined index and a scalar
I am trying to build out a docker container to do development in (keep breaking my env by accident).
Here is the compose file:
FROM ubuntu:20.04
USER root
RUN apt update -y && apt upgrade -y && \
apt install -y software-properties-common && \
add-apt-repository ppa:ubuntugis/ppa && \
apt update -y && apt install -y \
gdal-bin \
geotiff-bin \
git \
libgdal-dev \
libgl1 \
libspatialindex-dev \
wget \
python-is-python3 \
pip \
g++
RUN pip install Cython numpy
# RUN pip-compile requirements.txt
RUN git clone https://github.com/jgrss/geowombat.git
WORKDIR "/geowombat"
RUN pip install -r requirements.txt
RUN python setup.py install
RUN pip install sklearn-xarray pip-tools rtree ipykernel
# works but doesn't bind
RUN python -c "import geowombat as gw"
RUN python -c "import rasterio;from rasterio.crs import CRS; cc = CRS.from_epsg(4326)"
In the development environment I have always gotten the following error. I had been just commenting out the import, but that is stupid.
Any idea what is going on? I think this is an cython issue, but not sure how to resolve it.
Step 10/11 : RUN python -c "import geowombat as gw"
---> Running in 97cdbb6f6a2d
Traceback (most recent call last):
File "<string>", line 1, in <module>
File "/geowombat/geowombat/__init__.py", line 2, in <module>
from .core.api import open
File "/geowombat/geowombat/core/__init__.py", line 1, in <module>
from ..backends import transform_crs
File "/geowombat/geowombat/backends/__init__.py", line 2, in <module>
from .xarray_ import concat, mosaic
File "/geowombat/geowombat/backends/xarray_.py", line 7, in <module>
from ..core.windows import get_window_offsets
File "/geowombat/geowombat/core/windows.py", line 1, in <module>
from .util import n_rows_cols
File "/geowombat/geowombat/core/util.py", line 9, in <module>
from ..moving import moving_window
File "/geowombat/geowombat/moving/__init__.py", line 1, in <module>
from ._moving import moving_window
ModuleNotFoundError: No module named 'geowombat.moving._moving'
The command '/bin/sh -c python -c "import geowombat as gw"' returned a non-zero code: 1
The terminal process "/usr/bin/bash '-c', 'docker build --pull --rm -f "docker_debug" -t dbug2 "."'" terminated with exit code: 1.
Xarray is dropping support for xarray.ufuncs. Instead, NumPy can be used directly on a DataArray. The geowombat.radiometry.sr and geowombat.radiometry.angles modules contain xarray.ufuncs that can be replaced with numpy.
Look into adding a co-registration module that uses the AROSICS algorithm.
Installing on Windows 10, I followed the Easiest Install instructions using conda. This led to repeated 'Solving environment: failed...'.
Then I tried the Detailed Install further down on the same page, which is slightly different and seems redundant.
As this doesn't actually include a command to install geowombat at the end, I tried doing this with conda (per the 'Easiest') with the same result.
In the same environment, I also tried with pip: pip install git+https://github.com/jgrss/geowombat.
This results in the fiona install failing with 'A GDAL API version must be specified.'
Following the third set of alternate installation instructions here, (again, slightly different) I get the same fiona error. I tried setting GDAL_VERSION as an environment variable as instructed, but this led to 'ERROR: Failed building wheel for fiona'. I also tried installing fiona with conda, which worked, but then the pip install failed with another wheel error.
Maybe a minor tweek to improve band math.
import geowombat as gw
from geowombat.data import rgbn_suba, rgbn_subb
with gw.config.update(sensor='rgbn', scale_factor=0.0001):
with gw.open([rgbn_suba, rgbn_subb],
band_names=['blue', 'green', 'red', 'nir'],
mosaic=True,
bounds_by='union') as ds_mos:
# calculate NDVI
ndvi = ds_mos.gw.ndvi()
print(ndvi)
returns
<xarray.DataArray (band: 1, y: 283, x: 449)>
dask.array<broadcast_to, shape=(1, 283, 449), dtype=float64, chunksize=(1, 64, 64), chunktype=numpy.ndarray>
Coordinates:
* y (y) float64 2.05e+06 2.05e+06 2.05e+06 ... 2.049e+06 2.049e+06
* x (x) float64 7.929e+05 7.929e+05 7.929e+05 ... 7.952e+05 7.952e+05
* band (band) <U4 'ndvi'
Attributes:
transform: (5.0, 0.0, 792925.0, 0.0, -5.0, 2050115.0)
crs: +init=epsg:32618
res: (5.0, 5.0)
is_tiled: 0
nodatavals: (0.0, 0.0, 0.0, 0.0)
scales: (1.0, 1.0, 1.0, 1.0)
offsets: (0.0, 0.0, 0.0, 0.0)
resampling: nearest
pre-scaling: 0.0001
sensor: rgbn
drange: (-1, 1)
Shouldn't attributes be updated to reflect a single band after running band math? For instance nodatavals: (0.0), scales: (1.0) etc? This can be problematic I think when going to write out NDVI and needing to reset attributes.
@mmann1123 What do you think about adding an open method using STAC tools? Something that simplifies search and read.
I started experimenting here.
I am thinking something like this, which just uses pystac_client and stackstac.
import geowombat as gw
from rasterio.enums import Resampling
data_l = gw.open_stac(
stac_catalog='microsoft',
bounds='map.geojson',
collections=['landsat_c2_l2'],
bands=['red', 'green', 'blue', 'qa_pixel'],
mask_data=True,
resolution=100,
resampling=Resampling.cubic,
)
data_s2 = gw.open_stac(
stac_catalog='microsoft',
bounds='map.geojson',
collections=['sentinel_s2_l2a'],
bands=['B04', 'B03', 'B02'],
resolution=100,
resampling=Resampling.cubic,
epsg=32621
)print(data_l)
<xarray.DataArray 'stackstac-b46c4b9f62edc1d4b9cffdb0a334996a' (time: 4,
band: 3,
y: 269, x: 333)>
dask.array<fetch_raster_window, shape=(4, 3, 269, 333), dtype=float64, chunksize=(1, 1, 256, 256), chunktype=numpy.ndarray>
Coordinates: (12/27)
* time (time) datetime64[ns] 2020-07-04T13:12:45.30...
id (time) <U31 'LE07_L2SP_225078_20200704_02_T1...
* band (band) <U5 'red' 'green' 'blue'
* x (x) float64 6.626e+05 6.626e+05 ... 6.725e+05
* y (y) float64 -2.832e+06 -2.832e+06 ... -2.84e+06
landsat:wrs_path (time) <U3 '225' '225' '224' '224'
... ...
platform (time) <U9 'landsat-7' ... 'landsat-7'
eo:cloud_cover (time) float64 28.0 17.0 20.49 16.0
gsd int64 30
title (band) <U10 'Red Band' 'Green Band' 'Blue Band'
raster:bands object {'scale': 2.75e-05, 'nodata': 0, 'off...
epsg int64 32621
Attributes:
spec: RasterSpec(epsg=32621, bounds=(662580.0, -2840370.0, 672570....
crs: epsg:32621
transform: | 30.00, 0.00, 662580.00|\n| 0.00,-30.00,-2832300.00|\n| 0.0...
resolution: 30.0print(data.gw)
<geowombat.core.geoxarray.GeoWombatAccessor at 0x7f6c94740490>gw.merge_stac(data_l, data_s2)
<xarray.DataArray 'landsat_c2_l2+sentinel_s2_l2a' (time: 12, band: 3, y: 81,
x: 101)>
dask.array<getitem, shape=(12, 3, 81, 101), dtype=float64, chunksize=(1, 1, 81, 101), chunktype=numpy.ndarray>
Coordinates:
* time (time) datetime64[ns] 2020-07-04T13:12:45.307603 ... 2020-08-01T...
* band (band) <U8 'red' 'green' 'blue'
* y (y) float64 -2.832e+06 -2.832e+06 ... -2.84e+06 -2.84e+06
* x (x) float64 6.625e+05 6.626e+05 6.627e+05 ... 6.724e+05 6.725e+05
Attributes:
spec: RasterSpec(epsg=32621, bounds=(662500, -2840400, 672600, -28...
crs: epsg:32621
transform: | 100.00, 0.00, 662500.00|\n| 0.00,-100.00,-2832300.00|\n| 0...
resolution: 100
res: (100, 100)
collection: NoneThis might be my lack of understanding, but n_chunks seems to create problems when writing with n_worker and n_threads set.
with gw.open(l8_224078_20200518) as src:
# Write the data to a GeoTiff
src.sel(band=[3, 2, 1]).gw.to_raster('./output/output.tif',
n_workers=4, n_threads=2, n_chunks=16,
overwrite=True) fig, ax = plt.subplots(dpi=200)
with gw.open('./output/output.tif') as src:
src.where(src != 0).gw.imshow(robust=True, ax=ax)
plt.tight_layout(pad=1)I tried plotting the resulting .tif file in ArcMap and first, the predicted classes were not up to the number of classes in my training data. Second, I wasn't able to build unique values for the raster and the black bands around the raster are not disappearing when I set to display nodata value with no colour.
I think the major hurdle for most people (nubes like me at least) have with geowombat is the complexity of the install (outside linux). Someone already got a semi-working conda feedstock for v 1.2 up. But it points to .tar.gz release assets like this example from rasterio https://github.com/rasterio/rasterio/releases.
Maybe its because I am not the owner of the repository, but I'm not seeing the "create a release" button for geowombat on github for example. I am assuming this is something we can automate through circleci, but not sure.
Just wondering if that's in the works. Or if you can do it manually. I would love to get the conda feedstock running across all the systems.
Just wondering if it would be possible to allow both opening multiple images using stack_dim and at the same time subsetting using window.
Here's what I'd like to do:
import geowombat as gw
from rasterio.windows import Window
w = Window(row_off=0, col_off=0, height=100, width=100)
with gw.open([rgbn, rgbn],
band_names=['b1', 'g1', 'r1', 'n1', 'b2', 'g2', 'r2', 'n2'],
stack_dim='band',
window = w) as ds:
print(ds)gives an error "The band names do not match the output dimensions." Checking the error, it seems that the data shape is (2, 4, 100, 100). So,
with gw.open([rgbn, rgbn],
band_names=['b', 'g', 'r', 'n'],
time_names = ['old', 'new'],
window = w) as ds:
print(ds)works fine.
I'm using geowombat version 1.2.9
@mmann1123 I added a page with some editing examples.
Is that what you had in mind?
Add spatial-temporal fusion methods, such as StarFM.
Its late and this is a placeholder, but I think I found a problem with to_raster with separate = True (unless Im missing something).
ds.gw.to_raster('./landuse_stack_2019.tif',
n_workers=1, n_threads=12,bigtiff=True, compress='lzw',
separate=True, readxsize=3341, readysize= 2586, out_block_type='gtiff' )
2010
100%|███ | 4/4 [00:32<00:00, 8.17s/it]
Traceback (most recent call last):
File "<stdin>", line 16, in <module>
File "/home/mmann1123/Documents/github/geowombat/geowombat/core/geoxarray.py", line 719, in to_raster
**kwargs)
File "/home/mmann1123/Documents/github/geowombat/geowombat/core/io.py", line 884, in to_raster
group_keys = list(root.group_keys())
AttributeError: 'NoneType' object has no attribute 'group_keys'
>>>
Just looking to plot a tcap band and got an unexpected error.
fig, ax = plt.subplots(dpi=150)
with gw.config.update(sensor='qb', scale_factor=0.0001):
with gw.open(rgbn) as ds:
tcap = ds.gw.tasseled_cap()
tcap.sel(band='wetness').plot(robust=True, ax=ax)
plt.tight_layout(pad=1)throws
TypeError: Plotting requires coordinates to be numeric, boolean, or dates of type numpy.datetime64, datetime.datetime, cftime.datetime or pandas.Interval. Received data of type object instead.
Resolved in PR #87
Code
Current in conversion.py
def polygon_to_array(
self,
polygon,
col=None,
data=None,
...
dtype="uint16",
):
....
if col:
shapes = (
(geom, value) for geom, value in zip(dataframe.geometry, dataframe[col])
)
else:
shapes = dataframe.geometry.values
varray = rasterize(
shapes,
out_shape=(dst_height, dst_width),
transform=dst_transform,
fill=fill,
default_value=default_value,
all_touched=all_touched,
dtype=dtype,
)Problem
Solution
if col:
shapes = (
(geom, value) for geom, value in zip(dataframe.geometry, dataframe[col])
)
dtype=get_minimum_dtype(dataframe[col])
else:
shapes = dataframe.geometry.values
varray = rasterize(
shapes,
out_shape=(dst_height, dst_width),
transform=dst_transform,
fill=fill,
default_value=default_value,
all_touched=all_touched,
dtype=dtype,
)I was just mucking around with docker and testing the latest ubuntu version 22.4, and conda install.
I got the following errors on testing. Sops is installed, so not sure what's going on here.
I am wondering if it has to do more with conda install rather than the new ubuntu version
======================================================================
ERROR: test_coreg_data (test_coreg.TestCOREG)
----------------------------------------------------------------------
Traceback (most recent call last):
File "/geowombat/tests/test_coreg.py", line 29, in test_coreg_data
data = gw.coregister(
File "/root/miniconda3/lib/python3.9/site-packages/geowombat/core/sops.py", line 1242, in coregister
raise NameError
NameError
======================================================================
ERROR: test_coreg_shift (test_coreg.TestCOREG)
Tests a 1-pixel shift.
----------------------------------------------------------------------
Traceback (most recent call last):
File "/geowombat/tests/test_coreg.py", line 79, in test_coreg_shift
shifted = gw.coregister(
File "/root/miniconda3/lib/python3.9/site-packages/geowombat/core/sops.py", line 1242, in coregister
raise NameError
NameError
======================================================================
ERROR: test_coreg_transform_data (test_coreg.TestCOREG)
----------------------------------------------------------------------
Traceback (most recent call last):
File "/geowombat/tests/test_coreg.py", line 50, in test_coreg_transform_data
data = gw.coregister(
File "/root/miniconda3/lib/python3.9/site-packages/geowombat/core/sops.py", line 1242, in coregister
raise NameError
NameError
======================================================================
ERROR: test_array_type (test_properties.TestProperties)
----------------------------------------------------------------------
Traceback (most recent call last):
File "/geowombat/tests/test_properties.py", line 36, in test_array_type
self.assertTrue(src.gw.array_is_dask)
AttributeError: 'GeoWombatAccessor' object has no attribute 'array_is_dask'
======================================================================
ERROR: test_chunksize_check (test_properties.TestProperties)
----------------------------------------------------------------------
Traceback (most recent call last):
File "/geowombat/tests/test_properties.py", line 42, in test_chunksize_check
self.assertEqual(src.gw.check_chunksize(64, 128), 64)
AttributeError: 'GeoWombatAccessor' object has no attribute 'check_chunksize'
----------------------------------------------------------------------
When geowombat.open() is used within a configuration context with reference bounds set, the underlying rasterio.vrt.WarpedVRT() modifies the data. The issue appears to only occur with floating point data. Tests on thematic data did not result in incorrect data reads.
bounds = (left, bottom, right, top)
with gw.config.update(ref_bounds=bounds):
with gw.open(image) as src:
# do something hereresults in the correct positional read (i.e., the correct bounding box is read), but the data values are modified in rasterio.vrt.WarpedVRT().
I have run into this a few times now since the latest update. I haven't had time to look into it. Sorry with Fiona pulling at me all the time, I have about 1 hr of productive work time a day :(
You probably already have it resolved but thought I would check
import geowombat as gw
from geowombat.data import rgbn
from rasterio.windows import Window
w = Window(row_off=0, col_off=0, height=100, width=100)
with gw.open(rgbn,
band_names=['blue', 'green', 'red'],
num_workers=8,
indexes=[1, 2, 3],
window=w,
) as src:
print(src)---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
~/Documents/github/pyGIS/pygis/docs/test.py in
1185 num_workers=8,
1186 indexes=[1, 2, 3],
---> 1187 window=w,
1188 ) as src:
1189 print(src)
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/geowombat/core/api.py in __init__(self, filename, band_names, time_names, stack_dim, bounds, bounds_by, resampling, persist_filenames, netcdf_vars, mosaic, overlap, nodata, dtype, num_workers, **kwargs)
412 chunks=chunks,
413 num_workers=num_workers,
--> 414 **kwargs)
415
416 self.__filenames = [filename]
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/geowombat/core/api.py in read(filename, band_names, time_names, bounds, chunks, num_workers, **kwargs)
174 kwargs['window'] = from_bounds(*bounds, transform=src.gw.transform)
175
--> 176 ycoords, xcoords, attrs = get_attrs(src, **kwargs)
177
178 data = dask.compute(read_delayed(filename,
~/anaconda3/envs/pygisbookgw/lib/python3.7/site-packages/geowombat/core/api.py in get_attrs(src, **kwargs)
71 attrs = dict()
72
---> 73 attrs['transform'] = src.gw.transform
74
75 if hasattr(src, 'crs'):
AttributeError: 'DatasetReader' object has no attribute 'gw'
For a simple gpkg file, getting a key error. Wondering if the index should be used for the id?
sheds
Out[21]:
geometry
0 POLYGON ((37.28166 11.48168, 37.30379 11.48181...
1 POLYGON ((36.75196 11.33893, 36.78143 11.33915...
2 POLYGON ((37.35700 12.48734, 37.39401 12.48757...
3 POLYGON ((37.34259 12.49546, 37.37960 12.49569...
4 POLYGON ((36.89493 11.42643, 36.91459 11.42657...
.. ...
Extracting data using the following:
with gw.open('./data/NDVI_MODIS/NDVI_2013_01_01_250m.tif') as modis:
print(modis)
df = gw.extract(modis,'./data/CommunitySurvey/watershed_bboxes.gpkg',
#bands=3,
#band_names=['red'],
#frac=0.1,
#n_jobs=8,
#num_workers=8,
verbose=1)
Returns
Coordinates:
* band (band) int64 1
* y (y) float64 14.89 14.88 14.88 14.88 ... 3.412 3.41 3.408 3.406
* x (x) float64 33.01 33.01 33.01 33.01 ... 47.99 47.99 47.99 47.99
Attributes:
transform: (0.002245788210298804, 0.0, 33.00410353855122, 0.0, -0.002...
crs: +init=epsg:4326
res: (0.002245788210298804, 0.002245788210298804)
is_tiled: 1
nodatavals: (nan,)
scales: (1.0,)
offsets: (0.0,)
descriptions: ('NDVI',)
filename: ./data/NDVI_MODIS/NDVI_2013_01_01_250m.tif
resampling: nearest
0%| | 0/136 [00:00<?, ?it/s]
Traceback (most recent call last):
File "/home/mmann1123/Documents/github/IFPRI_ET_watersheds/scripts/extract_timeseries_to_poly.py", line 33, in <module>
verbose=1)
File "/home/mmann1123/anaconda3/envs/xr_fresh/lib/python3.7/site-packages/geowombat/core/sops.py", line 423, in extract
verbose=verbose)
File "/home/mmann1123/anaconda3/envs/xr_fresh/lib/python3.7/site-packages/geowombat/core/conversion.py", line 423, in prepare_points
n_jobs=n_jobs)
File "/home/mmann1123/anaconda3/envs/xr_fresh/lib/python3.7/site-packages/geowombat/core/conversion.py", line 464, in polygons_to_points
point_df = sample_feature(dfrow[id_column],
File "/home/mmann1123/anaconda3/envs/xr_fresh/lib/python3.7/site-packages/pandas/core/series.py", line 871, in __getitem__
result = self.index.get_value(self, key)
File "/home/mmann1123/anaconda3/envs/xr_fresh/lib/python3.7/site-packages/pandas/core/indexes/base.py", line 4404, in get_value
return self._engine.get_value(s, k, tz=getattr(series.dtype, "tz", None))
File "pandas/_libs/index.pyx", line 80, in pandas._libs.index.IndexEngine.get_value
File "pandas/_libs/index.pyx", line 90, in pandas._libs.index.IndexEngine.get_value
File "pandas/_libs/index.pyx", line 138, in pandas._libs.index.IndexEngine.get_loc
File "pandas/_libs/hashtable_class_helper.pxi", line 1619, in pandas._libs.hashtable.PyObjectHashTable.get_item
File "pandas/_libs/hashtable_class_helper.pxi", line 1627, in pandas._libs.hashtable.PyObjectHashTable.get_item
KeyError: 'id'
A declarative, efficient, and flexible JavaScript library for building user interfaces.
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
An Open Source Machine Learning Framework for Everyone
The Web framework for perfectionists with deadlines.
A PHP framework for web artisans
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
Some thing interesting about web. New door for the world.
A server is a program made to process requests and deliver data to clients.
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
Some thing interesting about visualization, use data art
Some thing interesting about game, make everyone happy.
We are working to build community through open source technology. NB: members must have two-factor auth.
Open source projects and samples from Microsoft.
Google ❤️ Open Source for everyone.
Alibaba Open Source for everyone
Data-Driven Documents codes.
China tencent open source team.