Comments (3)
Hi,
Thanks for trying out our new version. Two things I can think of:
- By
readRDS(), to my experience, the object is indeed loaded into the environment but just not shown in the "environment" panel of your RStudio. If you try operators like:
obj <- readRDS("oldfile.rds")
obj@norm.dataIt should show you a list of normalized matrices. and similar for the [email protected]
- However, force doing those above should probably keep spamming error messages on your screen. You can downgrade the package back to version 1.0.1 and normally load it, and have checks on the colnames and rownames of the matrices. In rliger2.0.0, we added strict checkpoints for cell and feature consistency to prevent potentially erroneous operations. What you will want to look at is, regard the row/colnames of the
[email protected]as the baseline, see if those innorm.dataare identical.scale.datashould be feature subset and transposed (cell x gene) in the old version, and you can see if the features are available inrownames([email protected][["datasetName"]]).
Above is kind of the overall idea of what should be taken care of, please feel free to reach out if you still see a problem.
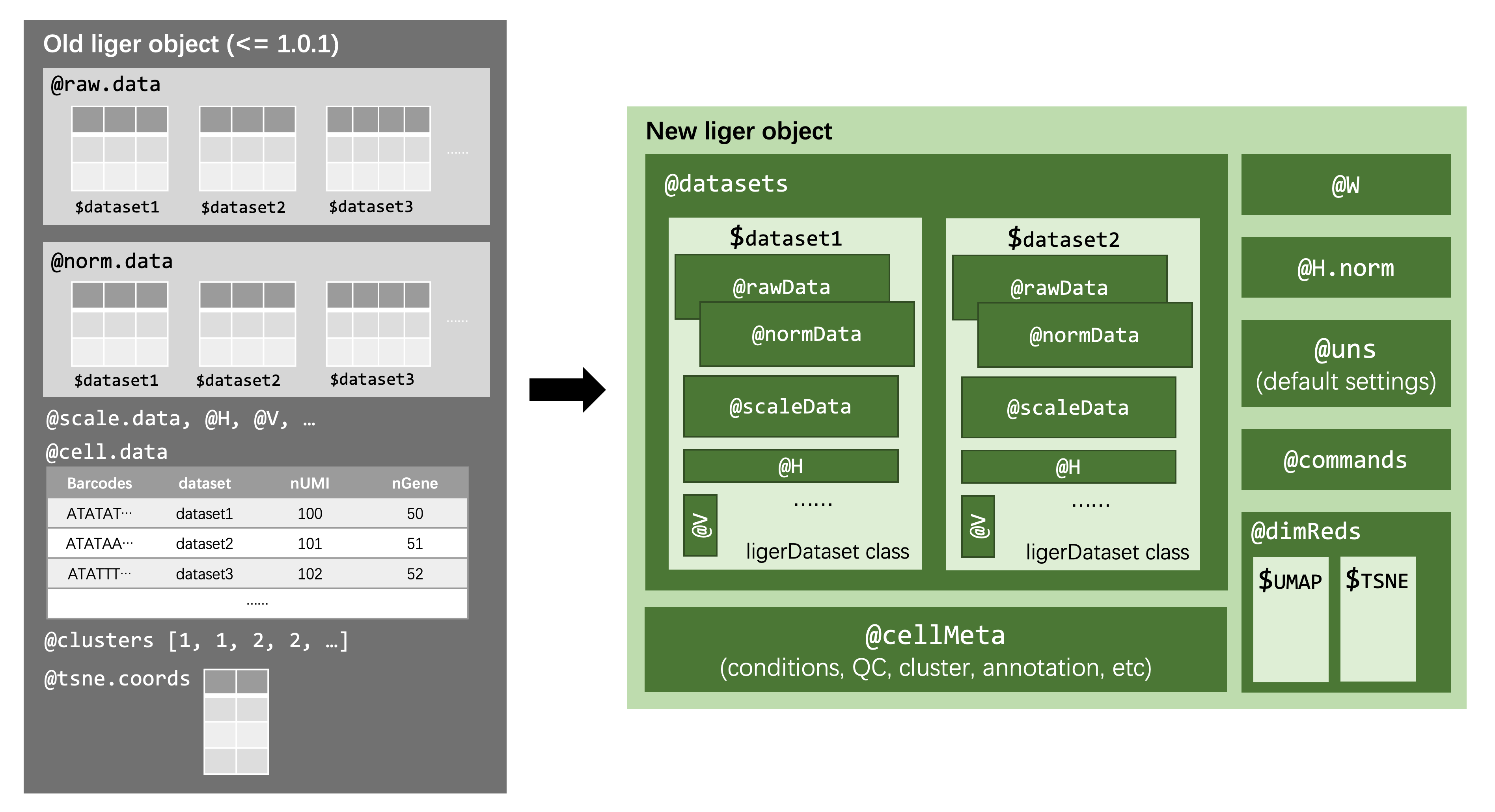
Here's a figure on the new documentation website that roughly shows how the structure of a liger object is refactored:
Let's keep this Issue open for people coming for rliger2 structure issues.
Appreciate it!
Yichen
from liger.
Thank you very much for the quick response and helpful resources! I will try that and add an update here with the outcome.
UDPATE: I was able to easily generate the new liger object by first reading in the old liger object as before and then manually working through the steps in the convertOldLiger function. (However, upon further inspection of the original object, I discovered that it does not include some critical metadata for my intended analysis, so I will have to stop at this point).
from liger.
Nice to hear that! By metadata did you mean some variables in [email protected]? You can use cellMeta<- method to manually insert any variable since you've already constructed an object of the new version. If a variable is named, the names will also be checked. Note that a prefix like "{datasetName}_" might have been added to the object colnames so that might creat some barriers.
from liger.
Related Issues (20)
- "shared factor neighborhood" not used anymore as described in the paper? HOT 3
- log transform data first? HOT 1
- Another request for help regarding SCTransform HOT 1
- Generate gene and promoter bed files
- Problems with scaleNotCenter() HOT 2
- Prepare for upcoming Seurat v5 release HOT 1
- Bug in Seurat's wrapper function HOT 1
- cran package archived HOT 6
- NaNs during `SelectGenes()` HOT 1
- Error on makeFeatureMatrix HOT 1
- How to improving similarity between snRNA-seq and snATAC-seq nuclei, by tuning parameters like 'k', 'lambda'? HOT 4
- Dataset specificity plot
- Error in `signal_stage()` HOT 4
- invalid type "list" in R_matrix_as_sparse error HOT 3
- no slot name "defaultCluster" for this object of class "liger" HOT 1
- Query on building shared factor neighborhood graph HOT 2
- Error when performing the Wilcoxon test on the peak counts HOT 1
- Error in ImputeKNN
- How can I determine the similarity scores between clusters using Liger's UINMF? HOT 5
Recommend Projects
-
 React
React
A declarative, efficient, and flexible JavaScript library for building user interfaces.
-
Vue.js
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
-
 Typescript
Typescript
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
-
TensorFlow
An Open Source Machine Learning Framework for Everyone
-
Django
The Web framework for perfectionists with deadlines.
-
Laravel
A PHP framework for web artisans
-
D3
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
-
Recommend Topics
-
javascript
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
-
web
Some thing interesting about web. New door for the world.
-
server
A server is a program made to process requests and deliver data to clients.
-
Machine learning
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
-
Visualization
Some thing interesting about visualization, use data art
-
Game
Some thing interesting about game, make everyone happy.
Recommend Org
-
Facebook
We are working to build community through open source technology. NB: members must have two-factor auth.
-
Microsoft
Open source projects and samples from Microsoft.
-
Google
Google ❤️ Open Source for everyone.
-
Alibaba
Alibaba Open Source for everyone
-
D3
Data-Driven Documents codes.
-
Tencent
China tencent open source team.

from liger.