pykeen / pykeen Goto Github PK
View Code? Open in Web Editor NEW🤖 A Python library for learning and evaluating knowledge graph embeddings
Home Page: https://pykeen.readthedocs.io/en/stable/
License: MIT License
🤖 A Python library for learning and evaluating knowledge graph embeddings
Home Page: https://pykeen.readthedocs.io/en/stable/
License: MIT License
Describe the bug
After training a model on fb15k237 dataset, I tried running a test with predict_tails.
When using this function, I had a KeyError resulting from calling predict_tails, this is because the predict function calls this line head_id = self.triples_factory.entity_to_id[head_label] which is fetching the dictionary entry for the entity, id pair.
This happened for /m/05hyfin the test set of fb15k237 dataset after training and just calling the predict function. Should a keyerror happen if the key is not found?
To Reproduce
Steps to reproduce the behavior:
from pykeen.triples.triples_factory import TriplesFactory
import pandas as pd
import torch
from pykeen.pipeline import pipeline
from numpy import asarray
from numpy import savetxt
import numpy as np
def main():
result = pipeline(
dataset= "fb15k237",
model="DistMult",
model_kwargs=dict(embedding_dim= 100),
regularizer= "no",
optimizer= "Adam",
optimizer_kwargs=dict(lr=0.001),
negative_sampler= "basic",
negative_sampler_kwargs=dict(
num_negs_per_pos= 500
),
training_kwargs=dict(
num_epochs=1, # just to be fast
batch_size=256,
),
)
model = result.model tf= TriplesFactory(path='test.txt') #this is the file from https://github.com/ZhenfengLei/KGDatasets/tree/master/FB15k-237
triples = tf.triples
results = []
model.predict_tails("/m/05hyf", "/film/film_subject/films") #KeyError hereExpected behavior
Runs and results in a dataframe of predictions.
Environment (please complete the following information):
Describe the bug
I am trying to use pykeen on my own data and following the BYOD section of the docs I am doing the following:
from pykeen.triples import TriplesFactory
from pykeen.pipeline import pipeline
tf = TriplesFactory(path="/tmp/exampledata.txt")
training, testing = tf.split()
pipeline_result = pipeline(
training_triples_factory=training, testing_triples_factory=testing, model="TransH"
)
Unfortunately I get the following:
Using random_state=1333753659 to split TriplesFactory(path="/tmp/out")
No random seed is specified. Setting to 2098687070.
No cuda devices were available. The model runs on CPU
Training epochs on cpu: 0%| | 0/5 [00:00<?, ?epoch/s]INFO:pykeen.training.training_loop:using stopper: <pykeen.stoppers.stopper.NopStopper object at 0x7f42e4842cc0>
Training epochs on cpu: 0%| | 0/5 [00:00<?, ?epoch/s]
Traceback (most recent call last):
File "test.py", line 8, in <module>
training_triples_factory=training, testing_triples_factory=testing, model="TransH"
File "/home/dobraczka/.local/share/virtualenvs/embedding-transformers-I3i1Obsv/lib/python3.7/site-packages/pykeen/pipeline.py", line 815, in pipeline
**training_kwargs,
File "/home/dobraczka/.local/share/virtualenvs/embedding-transformers-I3i1Obsv/lib/python3.7/site-packages/pykeen/training/training_loop.py", line 190, in train
num_workers=num_workers,
File "/home/dobraczka/.local/share/virtualenvs/embedding-transformers-I3i1Obsv/lib/python3.7/site-packages/pykeen/training/training_loop.py", line 376, in _train
slice_size,
File "/home/dobraczka/.local/share/virtualenvs/embedding-transformers-I3i1Obsv/lib/python3.7/site-packages/pykeen/training/training_loop.py", line 438, in _forward_pass
slice_size=slice_size,
File "/home/dobraczka/.local/share/virtualenvs/embedding-transformers-I3i1Obsv/lib/python3.7/site-packages/pykeen/training/slcwa.py", line 101, in _process_batch
positive_scores = self.model.score_hrt(positive_batch)
File "/home/dobraczka/.local/share/virtualenvs/embedding-transformers-I3i1Obsv/lib/python3.7/site-packages/pykeen/models/unimodal/trans_h.py", line 134, in score_hrt
d_r = self.relation_embeddings(hrt_batch[:, 1])
File "/home/dobraczka/.local/share/virtualenvs/embedding-transformers-I3i1Obsv/lib/python3.7/site-packages/torch/nn/modules/module.py", line 550, in __call__
result = self.forward(*input, **kwargs)
File "/home/dobraczka/.local/share/virtualenvs/embedding-transformers-I3i1Obsv/lib/python3.7/site-packages/torch/nn/modules/sparse.py", line 114, in forward
self.norm_type, self.scale_grad_by_freq, self.sparse)
File "/home/dobraczka/.local/share/virtualenvs/embedding-transformers-I3i1Obsv/lib/python3.7/site-packages/torch/nn/functional.py", line 1724, in embedding
return torch.embedding(weight, input, padding_idx, scale_grad_by_freq, sparse)
IndexError: index out of range in self
Loading e.g. the kinships dataset this way (i.e. concatenating train.txt, test.txt, valid.txt into a single file to have the same setting) I do not get this error. I have attached the data, which is a simple three-column tsv file.
exampledata.txt
To Reproduce
Steps to reproduce the behavior:
Expected behavior
Pipeline runs without errors and returns result.
Environment (please complete the following information):
As of the PyTorch 1.6 release on July 28th, 2020, there is a native type for tensors of complex numbers (see: https://pytorch.org/docs/stable/complex_numbers.html)
From @mberr: Referencing the 1.7.1 release tracker here, in case they mention something about it: pytorch/pytorch#47622
It would be interesting to test updated implementations of models using complex tensors such as RotatE and ComplEx to see if we can make them more elegant using this new trick. #292 is a good solution, but still not native. #134 presents a solution where the tensors themselves inside the Embedding are assigned the complex dtype like in:
...
# initialize weight outside of torch.nn.Embedding to sneak in the dtype definitiion
_weight = torch.empty((num_embeddings, embedding_dim), dtype=dtype)
self._embeddings = torch.nn.Embedding(
num_embeddings=num_embeddings,
embedding_dim=embedding_dim,
_weight=_weight,
)
...Then, the math in ComplEx can be updated like in:
# old
(h_re, h_im), (r_re, r_im), (t_re, t_im) = [split_complex(x=x) for x in (h, r, t)]
# new
h_re, h_im = h.real, h.imag
r_re, r_im = r.real, r.imag
t_re, t_im = t.real, t.imagHowever, this update is blocked by a major issue - the torch.nn.functional.embedding function does not currently support automatic differentiation on complex tensors.
Traceback (most recent call last):
File "/usr/local/opt/[email protected]/Frameworks/Python.framework/Versions/3.8/lib/python3.8/unittest/case.py", line 60, in testPartExecutor
yield
File "/usr/local/opt/[email protected]/Frameworks/Python.framework/Versions/3.8/lib/python3.8/unittest/case.py", line 676, in run
self._callTestMethod(testMethod)
File "/usr/local/opt/[email protected]/Frameworks/Python.framework/Versions/3.8/lib/python3.8/unittest/case.py", line 633, in _callTestMethod
method()
File "/Users/cthoyt/dev/pykeen/tests/test_models.py", line 439, in test_score_r_with_score_hrt_equality
raise e
File "/Users/cthoyt/dev/pykeen/tests/test_models.py", line 431, in test_score_r_with_score_hrt_equality
scores_r = self.model.score_r(batch)
File "/Users/cthoyt/dev/pykeen/src/pykeen/models/base.py", line 993, in score_r
expanded_scores = self.score_hrt(hrt_batch=hrt_batch)
File "/Users/cthoyt/dev/pykeen/src/pykeen/models/unimodal/complex.py", line 151, in score_hrt
h = self.entity_embeddings(indices=hrt_batch[:, 0])
File "/Users/cthoyt/.virtualenvs/pykeen/lib/python3.8/site-packages/torch/nn/modules/module.py", line 727, in _call_impl
result = self.forward(*input, **kwargs)
File "/Users/cthoyt/dev/pykeen/src/pykeen/nn/emb.py", line 177, in forward
x = self._embeddings(indices)
File "/Users/cthoyt/.virtualenvs/pykeen/lib/python3.8/site-packages/torch/nn/modules/module.py", line 727, in _call_impl
result = self.forward(*input, **kwargs)
File "/Users/cthoyt/.virtualenvs/pykeen/lib/python3.8/site-packages/torch/nn/modules/sparse.py", line 124, in forward
return F.embedding(
File "/Users/cthoyt/.virtualenvs/pykeen/lib/python3.8/site-packages/torch/nn/functional.py", line 1852, in embedding
return torch.embedding(weight, input, padding_idx, scale_grad_by_freq, sparse)
RuntimeError: embedding does not support automatic differentiation for outputs with complex dtype.Mock mock mock mock
RotatE throws a cpu/gpu tensor mismatch error on the optimizer step when running on gpu
File "/home/wwymak/code_experiments/pykeen_expts/hello_world_transe.py", line 19, in <module>
training_kwargs=dict(num_epochs=100)
File "/home/wwymak/libraries/pykeen/src/pykeen/pipeline.py", line 722, in pipeline
**training_kwargs,
File "/home/wwymak/libraries/pykeen/src/pykeen/training/training_loop.py", line 190, in train
num_workers=num_workers,
File "/home/wwymak/libraries/pykeen/src/pykeen/training/training_loop.py", line 370, in _train
self.optimizer.step()
File "/home/wwymak/anaconda3/envs/deep_graph/lib/python3.7/site-packages/torch/optim/adagrad.py", line 96, in step
state['sum'].addcmul_(1, grad, grad)
RuntimeError: expected device cpu but got device cuda:0
To Reproduce
running this simple code snippet on a gpu machine
results = pipeline(
dataset=Kinships,
model='RotatE',
random_seed=1235,
training_kwargs=dict(num_epochs=100)
)
print(results)Environment (please complete the following information):
(this error also happens on colab)
matplotlib, seaborn and word_cloud are needed for some of the plotting functionality and it would be useful to list them as optional installs in the documentation.
If I select the RGCN model, the trials return None:
Optuna init() got an unexpected keyword argument 'base_model_cls' ...
Choices for a categorical distribution should be a tuple of None, bool, int, float and str for persistent storage but contains <function symmetric_edge_weights at 0x2aac1328bb90> which is of type function.
Is there something I didn't realize? With other models it works fine.
I got the result of TransE on WN18RR dataset as follows, with 100 epoch and default parameter settings.
mean_rank={'best': 7396.350889192887, 'worst': 7396.3558481532145, 'avg': 7396.353368673051}, mean_reciprocal_rank={'best': 0.11426725607718224, 'worst': 0.11426725590337292, 'avg': 0.1142672559902559}, hits_at_k={'best': {1: 0.0032489740082079343, 3: 0.19493844049247605, 5: 0.24811901504787962, 10: 0.2973666210670315}, 'worst': {1: 0.0032489740082079343, 3: 0.19493844049247605, 5: 0.24811901504787962, 10: 0.2973666210670315}, 'avg': {1: 0.0032489740082079343, 3: 0.19493844049247605, 5: 0.24811901504787962, 10: 0.2973666210670315}}, adjusted_mean_rank=0.36492311337297334.
it seems a bit lower than the result reported in the paper
What about adding a mode that moves all entities that won't get used in evaluation over to training?
I suppose you mean the triples containing entities which are not in evaluation_entity_whitelist? We could think about this, but it would change the split ratio of the subsets' number of triples.
Maybe we add this functionality in our splitting functionalities (and emit appropriate warnings)? Or we look at it from a different perspective: we first split all triples of interest by split_ratio. Then we extend the training set by additional triples. Practically we need to make sure that the ID-mapping stays consistent.
Originally posted by @mberr in #62 (comment)
On which paper(s) is your requested model based.
Low-Dimensional Hyperbolic Knowledge Graph Embeddings
Chami et al. (2021)
https://arxiv.org/abs/2005.00545
List the original and other existing implementations of this model
Official pytorch implementation: https://github.com/HazyResearch/KGEmb
Tensorflow implementation: https://github.com/tensorflow/neural-structured-learning/tree/master/research/kg_hyp_emb
Relevance of this model
Ability to help and additional context
Your background and ability to help with the implementation of the model.
On which paper(s) is your requested model based.
https://arxiv.org/abs/2006.04986
List the original and other existing implementations of this model
No link provided in the paper
Relevance of this model
This model is a logical successor of RotatE, ComplEx, etc. @mali-git can also probably the authors to help implement it in PyKEEN since they're in his group
Ability to help and additional context
Your background and ability to help with the implementation of the model.
Make Block and Bases Decomposition are individual modules so the R-GCN class is agnostic to the chosen relation matrix decomposition.
Move the PR from https://github.com/mali-git/POEM_develop/pull/529
I am having difficulty which format my KG data should be in to be used with pykeen. What is the structure that it should have ?
Describe the bug
for a model trained on gpu, when I do `model.predict_tails(...) it gives an error of
....
/usr/local/lib/python3.6/dist-packages/pykeen/models/base.py in _novel(self, h, r, t)
399 """Return if the triple is novel with respect to the training triples."""
400 triple = torch.tensor(data=[h, r, t], dtype=torch.long, device=self.device).view(1, 3)
--> 401 return (triple == self.triples_factory.mapped_triples).all(dim=1).any().item()
402
403 def predict_scores_all_relations(
/usr/local/lib/python3.6/dist-packages/torch/tensor.py in wrapped(*args, **kwargs)
26 def wrapped(*args, **kwargs):
27 try:
---> 28 return f(*args, **kwargs)
29 except TypeError:
30 return NotImplemented
RuntimeError: expected device cuda:0 but got device cpu
(not sure if the model should always be on the cpu? but if so would be nice to update the docs to point this out, since if I trained the model on gpu I should expect to be able to do inference on the gpu as well?)
To Reproduce
Steps to reproduce the behavior:
Expected behavior
there shouldn't be a device mismatch
Environment (please complete the following information):
Additional information
an example on colab here
I have created a toy example as follows:
Brussels locatedIn Belgium
Belgium partOf EU
EU hasCapital Brussels
As far as I understand, there should be a trivial embedding solution in two dimensions (the three entities form a triangle, with the three relations being their connecting vectors). The code I use for this is as follows:
results = pipeline(
training_triples_factory=tf,
testing_triples_factory=tf,
model = 'TransE',
model_kwargs=dict(embedding_dim=2),
random_seed=1,
device='cpu',
)However, the result looks kind of unexpected:
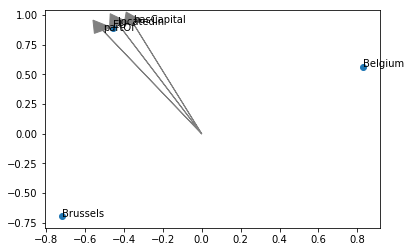
I also played with different random seeds and training parameters, but I never came to a sensible solution. DistMult and HolE, among others, come up with a similar solution.
Am I doing something wrong? Or is my conceptual understanding of the expected outcome incorrect?
As suggested, we're adding a list of projects that include Optuna to our docs. Can you confirm you're ok with this reference and the URL here? optuna/optuna#1605
Thanks!
You thought the 1.5 upgrade was confusing? Get ready for 2.0.0!
The release of v2.0 is approaching and we are going through the final details 🎯 Let us know if you need any last minute fixes or changes through this release candidate.
— OptunaAutoML (@OptunaAutoML) July 6, 2020
pip install optuna==2.0.0rc0https://t.co/4K8Oyc5a2K pic.twitter.com/CeL5JQ9jbv
Since #17 was merged before I got a chance to make a fuss about there not being documentation, it needs to be revisited.
RankBasedEvaluator.__init__() which should include explanations of what all parameters do (ks and filtered) as well as an explanation of integer hits@k versus fractional hits@k. References required.pipeline() and hpo_pipeline. Please provide unittests showing fractional hits@k in useBlocked by #18
Cross entropy loss and NSSA loss are the only two setwise loss formulations described in our paper. Its also a bit confusing becuase technically in the implementation, NSSA loss is inheriting from PairwiseLoss. Are there other setwise loss functions we could add, so there's more than 1?
Can you perhaps help me realize how I can use your framework properly? I tried following the documentation on how to train and evaluate embedding models in pykeen but after the OpenBioLink Dataset caused CUDA oom issues (partly because for most models slicing is not implemented or can I adapt any parameters to fit it?) (sidenote: how are the OBLF1 and F2 supposed to me used?) and I did not get the Neural Network implementations I was particularly interested in using to run I now tested some data set model combination that was actually used in your experiments and still the results seem far off anything you report (I am aware I did not run the same hyper parameter search -because different to your way where 24 hours get you through a good of parameters, for a training time of 107170 sec that was not feasible for a simple try out for me) the results are around hits@3 : 0.0005 and hits@10: 0.0007 for the attatched configuration. I have the 1.0.1-dev version installed and use Geforce GTX1080 2560 Cude-Kerne 11GB GDDR5 AERO GPUs.
from pykeen.pipeline import pipeline
pipeline_result = pipeline(
dataset='WN18RR',
model='RotatE',
model_kwargs=dict(
embedding_dim=500,
#automatic_memory_optimization=True,
),
training_kwargs=dict(num_epochs=1000),
optimizer='Adam',
optimizer_kwargs=dict(
lr=0.00008,
weight_decay=0.91,
),
training_loop='sLCWA',
loss='MarginRankingLoss',
evaluator='RankBasedEvaluator',
evaluator_kwargs=dict(filtered=True,),
negative_sampler="basic",
negative_sampler_kwargs=dict(num_negs_per_pos=1024)
)
print(pipeline_result)In #51 the novelty computation is introduced.
To enhance the computational efficiency and usability, automatic memory optimization as well as filtering has to be implemented.
Blocked by #18
The pointwise hinge loss sets the score of positive examples larger than a margin parameterλwhile reducing the scores of negative examples to values below−λ:
L(t_i,l_i) = max(0,λ−ˆl_i * f(ti))
where ˆl_i ∈ {−1,1}. The loss penalizes scores of positive examples which are smaller than λ, but does not impose any restriction on value s> λ. Similarly, negative scores larger than−λcontribute to the loss, whereas all values smaller than−λ do not have any loss contribution. Thereby, the model is not encouraged to further optimize triples which are already predicted well enough (according to the margin parameter λ).
Since each interaction model has a different range (e.g., the values of some interaction functions could be between [-inf,inf], some could be [0, inf], some could be [0, 1]), it makes the interpretation difficult. Even worse, this also has to do with interaction models' constrains on embeddings, etc.
It would be good for all interaction functions to define an "interpretation function" that remaps the results onto the range of [0,1]
From @mberr:
Obtaining such is an active research area and neither straight-forward, nor exists a "unique" solution. In case you are interested, you may want to look for keywords "model calibration" (e.g. Platt scaling), and for KG "triple classification".
needs step-by-step tutorial with screenshots
Give a directory and periodicity, save the model at the given state every x epochs into a new folder in the directory
Enable saving of models to AWS S3 using boto3. Will look very similar to the results from #28.
Add FTP saving to pipeline results and HPO results classes
Move from https://github.com/mali-git/POEM_develop/pull/478/files
Is your feature request related to a problem? Please describe.
I am trying to run experiments from a landmark. When the experiment runs, the results are config files and pkl file. There are files that contain MRR and Hits@1,3,10.
Describe the solution you'd like
I would like to see the predictions, not just the aggregate numerical results (like predictions for all triples). It is not clear how to do that from this point (especially after spending a lot of time training the model, then not being able to access the predictions).
Describe alternatives you've considered
Would it be possible to load from the pkl file and make predictions? I tried this but it did not work out. Please provide an example so this becomes more clear.
Additional context
Add any other context or screenshots about the feature request here.
Given an entity, its position (either head or tail), and a relation, return a ranked list of all entities and their scores. It should be clear if higher score or lower score means more likely to be a real edge. We might want to have it automatically filter out entities corresponding to edges already in the original knowledge graph.
This should be implemented in the top-level Model class, as all models should have this functionality.
Question: do models trained under LCWA or sLCWA have to behave differently here? Does this functionality have to behave differently based on the loss function?
Is your feature request related to a problem? Please describe.
I have trained some of the models so far by using the parameters in the experiments config files and some of them have taken an hour or less while others timed out after running on our university's supercomputer for over 23 hours. It's a little frustrating because I can't tell if something is wrong and I lose all the progress after training for a whole day.
Describe the solution you'd like
It would be great if we can have some estimates of how long the models would take so we know if something is wrong? Or maybe some hardware requirements to run them for that given time? I am running on one GPU and I can't tell if this timing is normal.
Describe alternatives you've considered
pykeen experiments command and also just taking the config file and using the parameters in the code by creating a pipeline and the experiments continue to take a long time.Additional context
Out of memory error screenshot here, triggered by running this code:
import torch map_location=torch.device('cpu') model = torch.load('trained_model.pkl') #distmult wn18 predictions_df = model.score_all_triples()
And also by running any predict all_triples experiment on the server I mentioned.

Blocked by #18
The pairwise logistic loss is defined as:
L(∆) = log(1 + exp(∆))
Thus, it can be seen as a soft-margin formulation of the pairwise hinge loss (MRL) with a margin of zero.
Currently, we support BCE and BCEAfterSigmoidLoss. However, training will fail when training a model with BCE because we do not provide the option predict_with_sigmoid in the scort_hrt/h/r/t() functions (we only support it in the predict_scores_all_heads/tails/relations() functions) .
We could remove the support for blank BCE, or we need to add the option predict_with_sigmoid in the scoring functions.
Enable the use of the HPO helper-functions for customized datasets.
Since all classes are now implemented directly in PyKEEN, we can move the default HPO ranges into the classes themselves rather than in the external dictionary
Thank you for the library. My usecase is to embed relations/entities in ConceptNet. The Conceptnet is not in RDF form, instead it is a csv file. So I need to convert a format compatible with PyKEEN.
I checked your source code and found this example Is this the data format for reading custom KGs?
Thanks in advance.
I'm interested in getting the resulting embeddings of entities and relations.
How to get this, please?
I couldn't find a method which could gives this.
It seems to me that the PipelineResult may have a method for this purpose?
Describe the bug
In the Bring Your Own Data section of the documentation, the results of a pipeline run are saved to the variable pipeline_result. Directly after result.save_to_directory('test_pre_stratified_transe') is called, triggering an error: NameError: name 'result' is not defined.
To Reproduce
Steps to reproduce the behavior:
NameError.Expected behavior
Rename result to pipeline_result (or vice versa) to prevent the NameError when copy pasting the example.
Environment (please complete the following information):
N/A. The issue is with the documentation.
Additional information
I looked for open or closed issues mentioning this typo and did not find them. Apologies if this typo is already known.
In the opposite scenario to #62/#83, we want to report several different sets of metrics based on each relation type. For example, for hetionet, you might want to specify edges or groups of edges to evaluate together:
from pykeen.pipeline import pipeline
pipeline_results = pipeline(
dataset='Hetionet',
model='RotatE',
evaluation_relation_whitelist=[
{'CtD', 'CpD'}, # evaluate the compund treats disease and compounds palliates disease relations together
'CcSE', # evaluate the compound causes side effect alone
{'CdG', 'CuG', 'CbG'}, # compound/gene regulations/bindings
{'GiG', 'GcG', 'Gr>G'}, # gene/gene relations,
None, # special entry None should group all remaining that haven't appeared in the rest
],
)None should be a special entry that groups everything you haven't considered. Maybe raise error if all have been covered and None is also added?all (or something else that can be written in a JSON configuration) should be a special entry that will do everything. Alternatively, "all" could always be reportedAlternate idea, using a dict for named subsets
from pykeen.pipeline import pipeline
pipeline_results = pipeline(
dataset='Hetionet',
model='RotatE',
evaluation_relation_whitelist={
'Compound-Disease': {'CtD', 'CpD'},
'Compound-SideEffect': 'CcSE',
'Compound-Gene': {'CdG', 'CuG', 'CbG'},
'Gene-Gene': {'GiG', 'GcG', 'Gr>G'},
'Remainder': None,
},
)It might be better overall to force usage of dictionaries so we can switch on the type (mapping vs. collection)
I just read through the pipeline and it seems like this is going to be quite a pain in the butt to do this...
When trying to use my own data I get the following error. I am not very experienced and don't know how to solve this or why this is happening.
ValueError Traceback (most recent call last)
<ipython-input-13-ef15ccdc9011> in <module>
3
4 tf = TriplesFactory(path=work_path + '/eucalyptus_triplets.txt')
----> 5 training, testing = tf.split()
6
7 pipeline_result = pipeline(
~\anaconda3\envs\pykeen\lib\site-packages\pykeen\triples\triples_factory.py in split(self, ratios, random_state, randomize_cleanup)
419 idx = np.arange(n_triples)
420 if random_state is None:
--> 421 random_state = np.random.randint(0, 2 ** 32 - 1)
422 logger.warning(f'Using random_state={random_state} to split {self}')
423 if isinstance(random_state, int):
mtrand.pyx in numpy.random.mtrand.RandomState.randint()
_bounded_integers.pyx in numpy.random._bounded_integers._rand_int32()
ValueError: high is out of bounds for int32Is your feature request related to a problem? Please describe.
The usage of Tensor.nonzero() (without arguments) has been deprecated, cf. pytorch/pytorch#40187
At least in
pykeen/src/pykeen/evaluation/evaluator.py
Line 360 in 630d872
we use the now deprecated version.
Describe the solution you'd like
Change all usages of nonzero to use to future-proof variant, i.e. in the above mentioned case
filter_batch = (entity_filter_test & relation_filter).nonzero(as_tuple=False) Describe alternatives you've considered
Keep it as it is. This might lead to problems once PyTorch removes the deprecated behaviour.
Additional context
Warning found in this notebook: https://gist.github.com/cthoyt/190233fd98a11306ceb13f2ee0e95a9e (scroll all the way to the bottom)
Right now, the margin and adversarial_temperature parameters do not have a default for NSSA loss. We need to pick reasonable ones and set it.
Issue #2 brought attention to the ConceptNet as a possible dataset to include with PyKEEN. They provide a tab-separated dump of the database here. It is not pre-stratified into training/testing/evaluation sets.
Because this file has additional columns besides head, relation, and tail, its inclusion will also require an updated to the SingleTabbedDataset such that the usecols keyword argument can be specified in the dataset's __init__()
Blocked by #196 because splitting algorithm is currently too slow for big datasets (with more than ~5 million triples)
We need to merge from the private development repository. It will take an awful lot dark magic to get to there:
[remote "poem"]
url = [email protected]:mali-git/POEM_develop.git
fetch = +refs/heads/*:refs/remotes/origin/*git pull poem master --allow-unrelated-histories --no-commit -S and pray to rebased god. Figured this out with lots of googling and eventually the perfect article, so thanks a bunch to the author @fdvBlocked by #18
The square error loss function computes the squared difference between the predicted scores and the labels l_i ∈ {0,1}:
L(t_i,l_i) = (1/2)(f(t_i)−l_i)^2
The squared error loss strongly penalizes predictions that deviate considerably from the labels, and is usually used for regression problems. For simple models it often permits more efficient optimization algorithms involving analytical solutions of sub-problems, e.g. the Alternating Least Squares algorithm
After I train the model on my own dataset, I got a file named trained_model.pkl ,how to load the saved model and use it to predict?
Currently, it is difficult to understand the data structure (i.e. what I am passing to the model).
It may be useful to add a description page to help in developing custom models and datasets and therefore, extend the framework.
Furthermore, I think that an example based on Graph Neural Networks (maybe with PyTorch Geometric) will be very appreciated.
I am trying to use the RGCN algorithm and run into the following error.
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-6-231451742934> in <module>
5 training, testing = tf.split()
6
----> 7 pipeline_result = pipeline(
8 training_triples_factory=training,
9 testing_triples_factory=testing,
~\anaconda3\envs\pykeen\lib\site-packages\pykeen\pipeline.py in pipeline(dataset, dataset_kwargs, training_triples_factory, testing_triples_factory, validation_triples_factory, evaluation_entity_whitelist, evaluation_relation_whitelist, model, model_kwargs, loss, loss_kwargs, regularizer, regularizer_kwargs, optimizer, optimizer_kwargs, clear_optimizer, training_loop, negative_sampler, negative_sampler_kwargs, training_kwargs, stopper, stopper_kwargs, evaluator, evaluator_kwargs, evaluation_kwargs, result_tracker, result_tracker_kwargs, metadata, device, random_seed, use_testing_data)
711
712 model = get_model_cls(model)
--> 713 model_instance: Model = model(
714 triples_factory=training_triples_factory,
715 **model_kwargs,
~\anaconda3\envs\pykeen\lib\site-packages\pykeen\models\unimodal\rgcn.py in __init__(self, triples_factory, embedding_dim, automatic_memory_optimization, loss, predict_with_sigmoid, preferred_device, random_seed, num_bases_or_blocks, num_layers, use_bias, use_batch_norm, activation_cls, activation_kwargs, base_model, sparse_messages_slcwa, edge_dropout, self_loop_dropout, edge_weighting, decomposition, buffer_messages)
231 num_bases_or_blocks = triples_factory.num_relations // 2 + 1
232 if num_bases_or_blocks > triples_factory.num_relations:
--> 233 raise ValueError('The number of bases should not exceed the number of relations.')
234 elif self.decomposition == 'block':
235 if num_bases_or_blocks is None:
ValueError: The number of bases should not exceed the number of relations.
I assumed that bases are the same as nodes. When changing the number of relation types to be more than the different number of nodes It seemed to work.
However I want to apply the RGCN approach to a graph with 30 000 different nodes and only 6 different relation types between the nodes. From what I understand about the mathematics behind the RGCN model this should be possible, why is it recommended then in the error to have so many different relation types?
I am also sorry if this is not the platform to ask questions like this, but it seems to be the only platform where I consistently get answers about this package, presumably due to how new it still is.
A declarative, efficient, and flexible JavaScript library for building user interfaces.
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
An Open Source Machine Learning Framework for Everyone
The Web framework for perfectionists with deadlines.
A PHP framework for web artisans
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
Some thing interesting about web. New door for the world.
A server is a program made to process requests and deliver data to clients.
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
Some thing interesting about visualization, use data art
Some thing interesting about game, make everyone happy.
We are working to build community through open source technology. NB: members must have two-factor auth.
Open source projects and samples from Microsoft.
Google ❤️ Open Source for everyone.
Alibaba Open Source for everyone
Data-Driven Documents codes.
China tencent open source team.