🔗 Website | 📃 Docs | ⚙️ Install Guide | 📝 Tutorial | 💡 Examples
Optuna is an automatic hyperparameter optimization software framework, particularly designed for machine learning. It features an imperative, define-by-run style user API. Thanks to our define-by-run API, the code written with Optuna enjoys high modularity, and the user of Optuna can dynamically construct the search spaces for the hyperparameters.
Optuna has modern functionalities as follows:
- Lightweight, versatile, and platform agnostic architecture
- Handle a wide variety of tasks with a simple installation that has few requirements.
- Pythonic search spaces
- Define search spaces using familiar Python syntax including conditionals and loops.
- Efficient optimization algorithms
- Adopt state-of-the-art algorithms for sampling hyperparameters and efficiently pruning unpromising trials.
- Easy parallelization
- Scale studies to tens or hundreds of workers with little or no changes to the code.
- Quick visualization
- Inspect optimization histories from a variety of plotting functions.
We use the terms study and trial as follows:
- Study: optimization based on an objective function
- Trial: a single execution of the objective function
Please refer to the sample code below. The goal of a study is to find out the optimal set of
hyperparameter values (e.g., regressor and svr_c) through multiple trials (e.g.,
n_trials=100). Optuna is a framework designed for automation and acceleration of
optimization studies.
Sample code with scikit-learn
import ...
# Define an objective function to be minimized.
def objective(trial):
# Invoke suggest methods of a Trial object to generate hyperparameters.
regressor_name = trial.suggest_categorical('regressor', ['SVR', 'RandomForest'])
if regressor_name == 'SVR':
svr_c = trial.suggest_float('svr_c', 1e-10, 1e10, log=True)
regressor_obj = sklearn.svm.SVR(C=svr_c)
else:
rf_max_depth = trial.suggest_int('rf_max_depth', 2, 32)
regressor_obj = sklearn.ensemble.RandomForestRegressor(max_depth=rf_max_depth)
X, y = sklearn.datasets.fetch_california_housing(return_X_y=True)
X_train, X_val, y_train, y_val = sklearn.model_selection.train_test_split(X, y, random_state=0)
regressor_obj.fit(X_train, y_train)
y_pred = regressor_obj.predict(X_val)
error = sklearn.metrics.mean_squared_error(y_val, y_pred)
return error # An objective value linked with the Trial object.
study = optuna.create_study() # Create a new study.
study.optimize(objective, n_trials=100) # Invoke optimization of the objective function.Note
More examples can be found in optuna/optuna-examples.
The examples cover diverse problem setups such as multi-objective optimization, constrained optimization, pruning, and distributed optimization.
Optuna is available at the Python Package Index and on Anaconda Cloud.
# PyPI
$ pip install optuna# Anaconda Cloud
$ conda install -c conda-forge optunaOptuna has integration features with various third-party libraries. Integrations can be found in optuna/optuna-integration and the document is available here.
Supported integration libraries
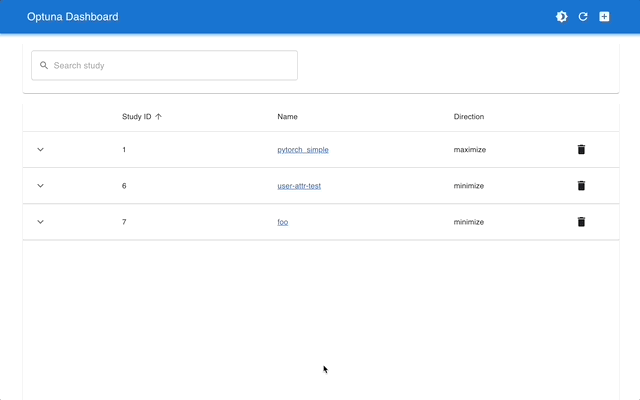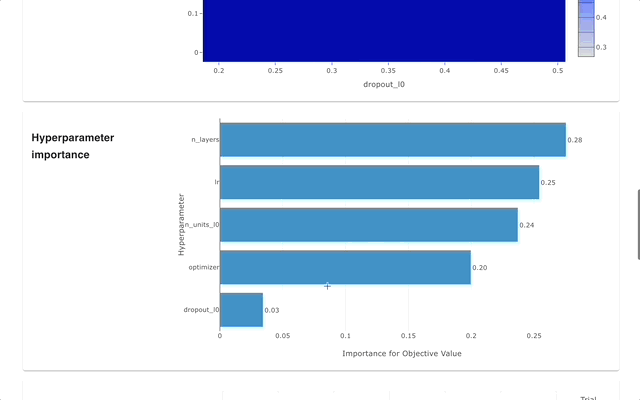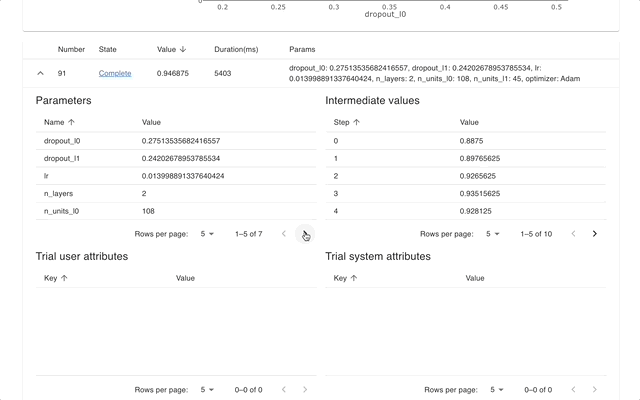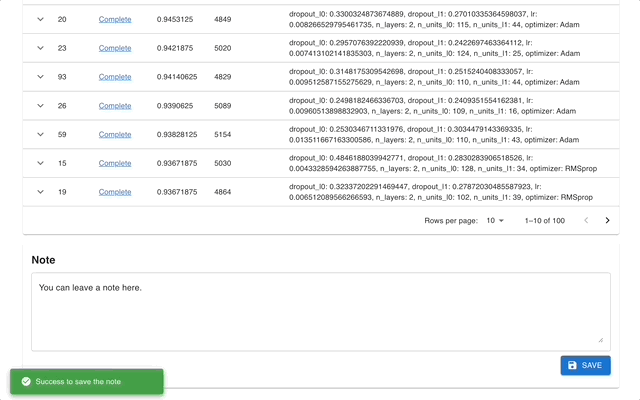
Optuna Dashboard is a real-time web dashboard for Optuna. You can check the optimization history, hyperparameter importance, etc. in graphs and tables. You don't need to create a Python script to call Optuna's visualization functions. Feature requests and bug reports are welcome!
optuna-dashboard can be installed via pip:
$ pip install optuna-dashboardTip
Please check out the convenience of Optuna Dashboard using the sample code below.
Sample code to launch Optuna Dashboard
Save the following code as optimize_toy.py.
import optuna
def objective(trial):
x1 = trial.suggest_float("x1", -100, 100)
x2 = trial.suggest_float("x2", -100, 100)
return x1 ** 2 + 0.01 * x2 ** 2
study = optuna.create_study(storage="sqlite:///db.sqlite3") # Create a new study with database.
study.optimize(objective, n_trials=100)Then try the commands below:
# Run the study specified above
$ python optimize_toy.py
# Launch the dashboard based on the storage `sqlite:///db.sqlite3`
$ optuna-dashboard sqlite:///db.sqlite3
...
Listening on http://localhost:8080/
Hit Ctrl-C to quit.- GitHub Discussions for questions.
- GitHub Issues for bug reports and feature requests.
Any contributions to Optuna are more than welcome!
If you are new to Optuna, please check the good first issues. They are relatively simple, well-defined, and often good starting points for you to get familiar with the contribution workflow and other developers.
If you already have contributed to Optuna, we recommend the other contribution-welcome issues.
For general guidelines on how to contribute to the project, take a look at CONTRIBUTING.md.
If you use Optuna in one of your research projects, please cite our KDD paper "Optuna: A Next-generation Hyperparameter Optimization Framework":
BibTeX
@inproceedings{akiba2019optuna,
title={{O}ptuna: A Next-Generation Hyperparameter Optimization Framework},
author={Akiba, Takuya and Sano, Shotaro and Yanase, Toshihiko and Ohta, Takeru and Koyama, Masanori},
booktitle={The 25th ACM SIGKDD International Conference on Knowledge Discovery \& Data Mining},
pages={2623--2631},
year={2019}
}