ggdist is an R package that provides
a flexible set of {ggplot2} geoms and stats designed especially for
visualizing distributions and uncertainty. It is designed for both
frequentist and Bayesian uncertainty visualization, taking the view that
uncertainty visualization can be unified through the perspective of
distribution visualization: for frequentist models, one visualizes
confidence distributions or bootstrap distributions (see
vignette("freq-uncertainty-vis")); for Bayesian models, one visualizes
probability distributions (see the
tidybayes package, which builds
on top of {ggdist}).
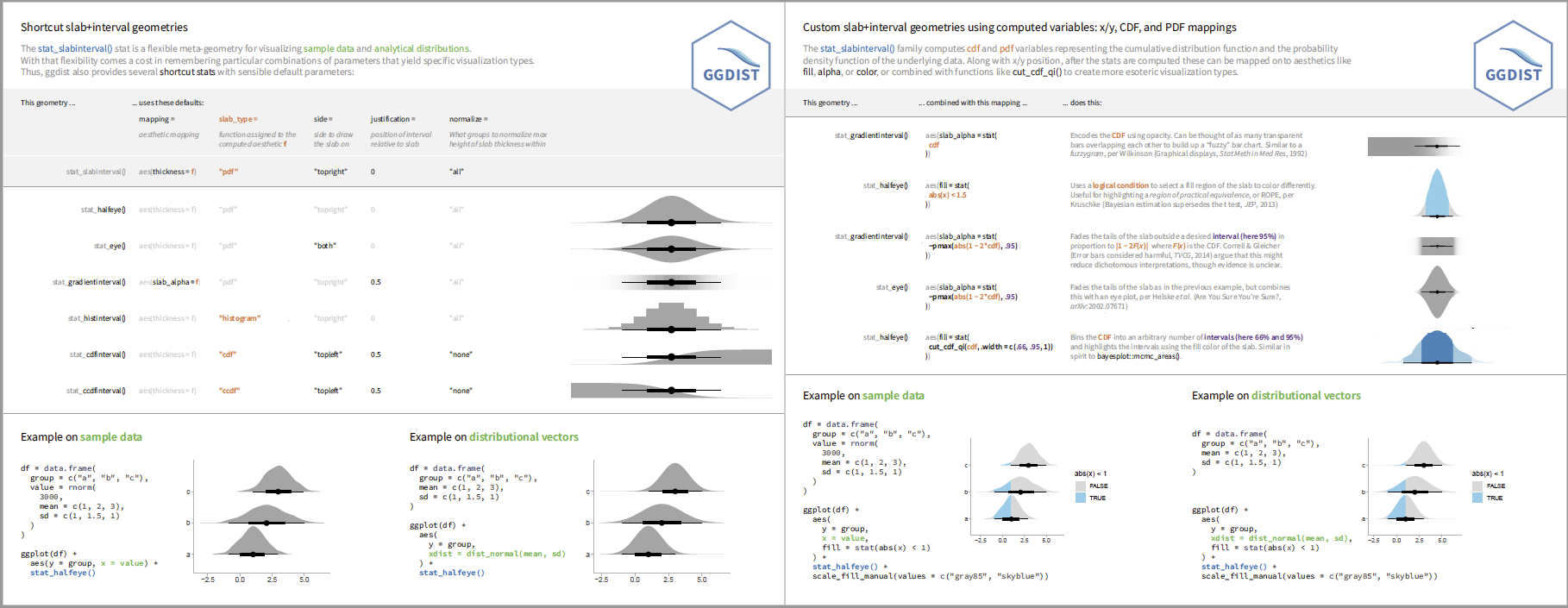
The geom_slabinterval() / stat_slabinterval() family (see
vignette("slabinterval")) makes it easy to visualize point summaries
and intervals, eye plots, half-eye plots, ridge plots, CCDF bar plots,
gradient plots, histograms, and more:
The geom_dotsinterval() / stat_dotsinterval() family (see
vignette("dotsinterval")) makes it easy to visualize dot+interval
plots, Wilkinson dotplots, beeswarm plots, and quantile dotplots (and
combined with half-eyes, composite plots like rain cloud plots):
The geom_lineribbon() / stat_lineribbon() family (see
vignette("lineribbon")) makes it easy to visualize fit lines with an
arbitrary number of uncertainty bands:
All stats in {ggdist} also support visualizing analytical
distributions and vectorized distribution data types like
distributional
objects or posterior::rvar() objects. This is particularly useful when
visualizing uncertainty in frequentist models (see
vignette("freq-uncertainty-vis")) or when visualizing priors in a
Bayesian analysis.
The {ggdist} geoms and stats also form a core part of the
tidybayes package (in fact, they
originally were part of {tidybayes}). For examples of the use of
{ggdist} geoms and stats for visualizing uncertainty in Bayesian
models, see the vignettes in {tidybayes}, such as
vignette("tidybayes", package = "tidybayes") or
vignette("tidy-brms", package = "tidybayes").
These cheat sheets focus on the slabinterval family of geometries:
You can install the currently-released version from CRAN with this R command:
install.packages("ggdist")Alternatively, you can install the latest development version from GitHub with these R commands:
install.packages("devtools")
devtools::install_github("mjskay/ggdist"){ggdist} aims to have minimal additional dependencies beyond those
already required by {ggplot2}. The {ggdist} dependencies fall into
the following categories:
-
{ggplot2}. -
Packages that
{ggplot2}also depends on. These packages add no additional dependency cost because{ggplot2}already requires them:{rlang},{cli},{scales},{tibble},{vctrs},{withr},{gtable}, and{glue}. -
Packages that
{ggplot2}does not depend on. These are all well-maintained packages with few dependencies and a clear need within{ggdist}:{distributional}: this implementation of distribution vectors powers much of{ggdist}. This package adds minimal additional cost, as its only dependency that is not also a dependency of{ggplot2}is{numDeriv}, which is needed by{ggdist}anyway (see below).{numDeriv}: used for calculating Jacobians of scale transformations. Needed because testing has revealed common situations wherestats::numericDeriv()fails but{numDeriv}does not. Widely used by other CRAN packages and has no additional dependencies.{quadprog}: Used to solve constrained optimization problems during different parts of dotplot layout, particularly to avoid dot overlaps in the"bin"and"weave"layouts. Widely used by other CRAN packages and has no additional dependencies.{Rcpp}: Used to implement faster dotplot layout. Widely used by other CRAN packages and has no additional dependencies.
I welcome feedback, suggestions, issues, and contributions! If you have
found a bug, please file it
here with minimal code to
reproduce the issue. Pull requests should be filed against the
dev branch. I am not
particularly reliable over email, though you can try to contact me at
[email protected]. A Twitter DM is
more likely to elicit a response.
Matthew Kay (2024). ggdist: Visualizations of Distributions and Uncertainty in the Grammar of Graphics. IEEE Transactions on Visualization and Computer Graphics, 30(1), 414–424. DOI: 10.1109/TVCG.2023.3327195.
Matthew Kay (2024). ggdist: Visualizations of Distributions and Uncertainty. R package version 3.3.2, https://mjskay.github.io/ggdist/. DOI: 10.5281/zenodo.3879620.