Update April 2023: no more daily updates.
Some counties in Germany have stopped reporting data in January 2023, and it is probably fair to say that by now there is not so much demand anymore for a project like this.
I would like to say a huge Thank You for your tremendous interest, for contributing, for critical discussion, and for helping reveal the most brittle kind of edge cases. All that helped keep the data flowing while rarely compromising on quality.
In the future, we hopefully do not need an underground software engineering effort like this anymore, and state of Germany will be able to expose relevant data quickly via useful and robust interfaces.
For me personally, this was a rather significant engineering effort and I learned a whole lot. I feel both, pain and happiness when I go through the almost 300 patches that I had been working on since March 20, 2020.
The following list is based on a non-exhaustive web search:
- Björn Thor Arnarson, 2021. How a school holiday led to persistent COVID-19 outbreaks in Europe. Nature Scientific Reports (11). HTML
- Luigi Palatella et al., 2021. A phenomenological estimate of the true scale of COVID-19 from primary data. Chaos, Solitons & Fractals (146). HTML
- Marcella Alsan et al., 2020; revised 2023. Civil Liberties in Times of Crisis. NBER Working Papers. HTML, PDF, Appendix PDF
- Naqvi A, 2021. COVID-19 European regional tracker. Scientific Data (8). HTML, PDF
- Philip Eisenlohr, 2020. Comparative visualization of global COVID-19 progression over time. medium.com. HTML
- Teodoro Alamo et al., 2020. COVID-19: Open-Data Resources for Monitoring, Modeling, and Forecasting the Epidemic. Electronics (9). HTML
- S Di Federico, 2022. Availability of open data related to COVID-19 epidemic in Italy. Annali di Igiene. HTML, PDF
- Mattia Mazzoli et al., 2021. Interplay between mobility, multi-seeding and lockdowns shapes COVID-19 local impact. PLOS Computational Biology. HTML
- Elaine Ford, Winfried Weck, 2020. Internet and the Pandemic in the Americas. The First Health Crisis of the Digital Era. Book. Konrad Adenauer Stiftung. HTML
- Björn Thor Arnarson, 2021. Breaks and Breakouts: Explaining the Persistence of COVID-19. SSRN Electronic Journal. HTML
- Abdollah Jalilian, Jorge Mateu, 2021. A hierarchical spatio-temporal model to analyze relative risk variations of COVID-19: a focus on Spain, Italy and Germany. Stochastic Environmental Research and Risk Assessment. HTML
- Ben-Hur Cardoso, Sebastian Gonçalves, 2020. Universal scaling law for COVID-19 propagation in urban centers. Preprint. HTML, PDF
- Fabrizio Pecoraro, Daniela Luzi, 2021. Open Data Resources on COVID-19 in Six European Countries: Issues and Opportunities. Int J Environ Res Public Health. HTML, PDF
- Teodoro Alamo et al, 2020. Data-Driven Methods to Monitor, Model, Forecast and Control Covid-19 Pandemic: Leveraging Data Science, Epidemiology and Control Theory. HTML, PDF
- J. Delgado, 2021. Applying the FAIR Principles to Accelerate Health Research in Europe in the Post COVID-19 Era. Proceedings of the 2021 EFMI Special Topic Conference. Google Books Preview
- Luis Baibás et al., 2020. COVID-19 effective reproductive ratio determination: An application, and analysis of issues and influential factors. Preprint. HTML
- https://www.researchgate.net/publication/358965720_Predict_Covid_Cases_with_LSTM-Network
- https://gist.github.com/denis-bz/d5259a341ba3b134fc2ecf2293b8f159
- https://data.world/liz-friedman/covid-19-in-germany
- https://www.cg.tuwien.ac.at/courses/Visualisierung2/HallOfFame/2020/projects/Goertler2018-Alexander%20Bayerl_44328754_assignsubmission_file_/doc/CodeDocumentation.pdf
- https://www.medrxiv.org/content/medrxiv/suppl/2020/04/07/2020.04.05.20054288.DC1/2020.04.05.20054288-1.pdf
- https://quantitas.it/data/coronavirus/
- https://www.studocu.com/de/document/technische-universitat-munchen/grundlagen-der-kunstlichen-intelligenz-in2062/exercise-4/21966548
- https://corona-open-data.ckan.de/dataset/covid-19-case-numbers-for-germany/resource/59f3eccd-0376-4b31-a1fa-498059c6176f
(see below for an English version)
- COVID-19 Fallzahlen für Bundesländer und Landkreise.
- Mehrfach täglich automatisiert aktualisiert.
- Mit Zeitreihen (inkl. 7-Tage-Inzidenz-Zeitreihen).
- Aktuelle Einwohnerzahlen und GeoJSON-Daten, mit transparenten Quellen.
- Präzise maschinenlesbare CSV-Dateien. Zeitstempel in ISO 8601-Notation, Spaltennamen nutzen u.a. ISO 3166 country codes.
- Zwei verschiedene Perspektiven:
- Die offiziellen Zeitreihen des RKI, auf Basis einer ArcGIS HTTP Schnittstelle (docs) des Esri COVID-19 GeoHub Deutschland. Diese Zeitreihen werden täglich in die Vergangenheit hinein aktualisiert und bieten einen kuratierten Blick auf die vergangenen Monate und Wochen.
- Die Zeitreihen der Risklayer GmbH-koordinierten Crowdsourcing-Initiative (die Datenbasis für tagesaktuelle Zahlen einiger deutscher Medien, z. B. des ZDF aber auch der JHU).
- Historical (time series) data for individual Bundesländer and Landkreise (states and counties).
- Automatic updates, multiple times per day.
- 7-day incidence time series (so that you don't need to compute those).
- Population data and GeoJSON data, with transparent references and code for reproduction.
- Provided through machine-readable (CSV) files: timestamps are encoded using ISO 8601 time string notation. Column names use the ISO 3166 notation for individual states.
- Two perspectives on the historical evolution:
- Official RKI time series data, based on an ArcGIS HTTP API (docs) provided by the Esri COVID-19 GeoHub Deutschland. These time series are being re-written as data gets better over time (accounting for delay in reporting etc), and provide a credible, curated view into the past weeks and months.
- Time series data provided by the Risklayer GmbH-coordinated crowdsourcing effort (the foundation for what various German newspapers and TV channels show on a daily basis, such as the ZDF but also the foundation for what the JHU) publishes about Germany.
You probably have a number of questions. Just as I had (and still have). Your feedback, your contributions, and your questions are highly appreciated! Please use the GitHub issue tracker (preferred) or contact me via mail. For updates, you can also follow me on Twitter: @gehrcke.
Note that these plots are updated multiple times per day. Feel free to hotlink them.
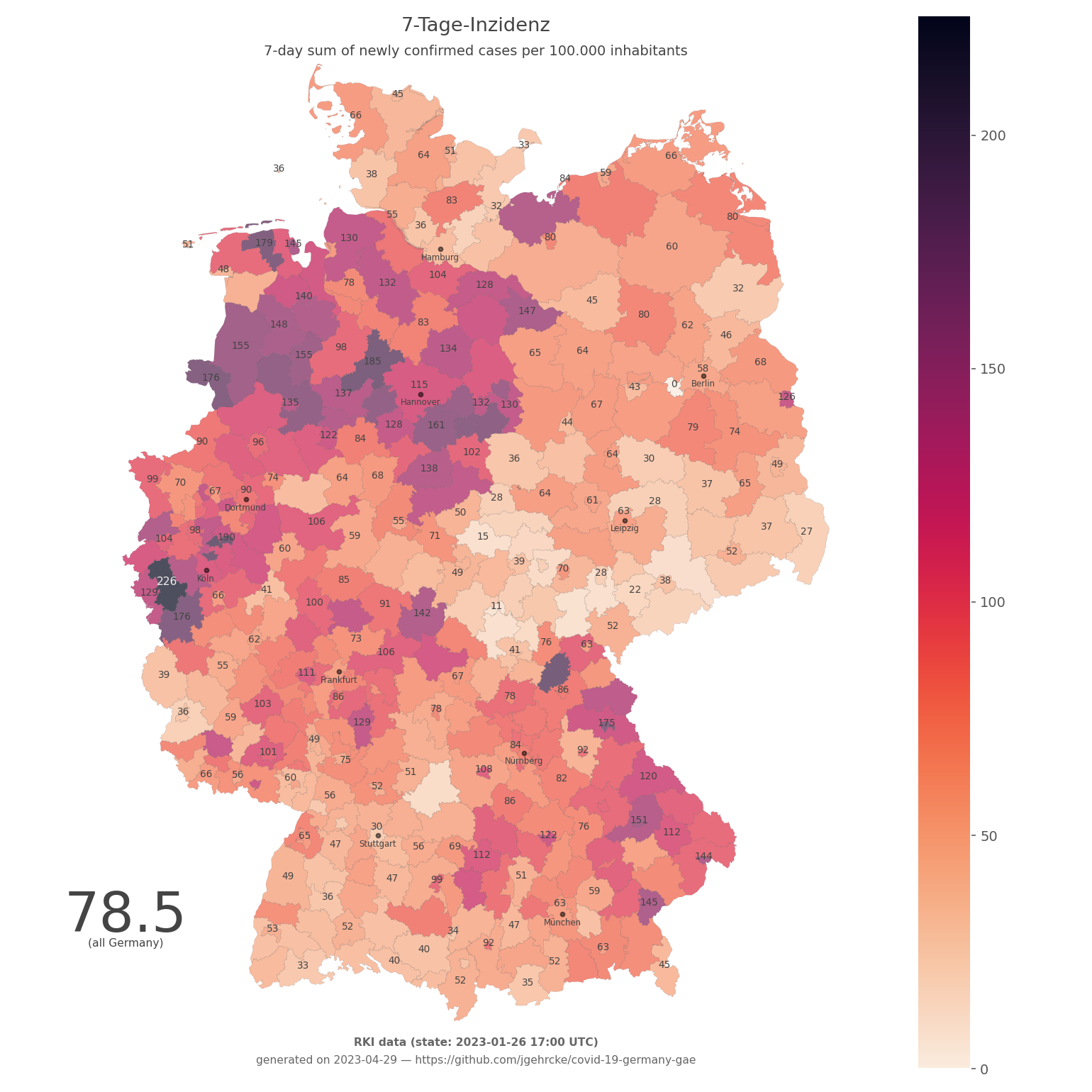
- germany-heatmap-7ti-rki.png (also available as PDF):
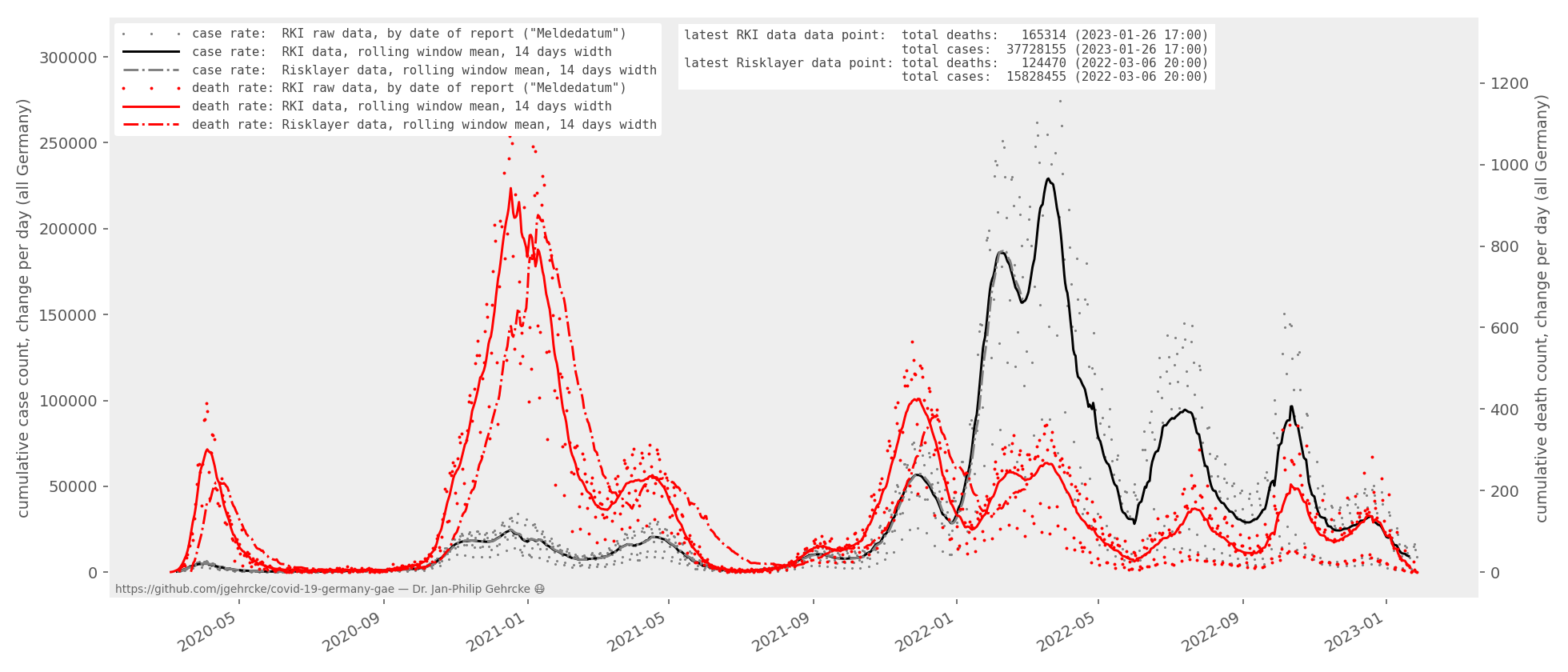
- daily-change-plot-latest.png (also available as PDF):
Note: there is a systematic difference between the RKI data-based death rate curve and the Risklayer-based death rate curve. Both curves are wrong, and yet both curves are legit. The incidents of death that we learn about today may have happened days or weeks in the past. Neither curve attempts to show the exact time of death (sadly! :-)) The RKI curve, in fact, is based on the point in time when each corresponding COVID-19 case that led to death was registered in the first place ("Meldedatum" of the corresponding case). The Risklayer data set to my knowledge pretends as if the incidents of death we learn about today happened yesterday. While this is not true, the resulting curve is a little more intuitive. Despite its limitations, the Risklayer data set is the best view on the "current" evolution of deaths that we have.
- RKI data (most credible view into the past): time series data provided by the Robert Koch-Institut (updated daily):
- cases-rki-by-ags.csv and deaths-rki-by-ags.csv: per-Landkreis time series
- cases-rki-by-state.csv and deaths-rki-by-state.csv: per-Bundesland time series
- 7-day incidence time series resolved by county based on RKI data can be found in
more-data/. - This is the only data source that rigorously accounts for Meldeverzug (reporting delay). The historical evolution of data points in these files is updated daily based on a (less accessible) RKI ArcGIS system. These time series see amendments weeks and months into the past as data gets better over time. This data source has its strength in the past, but it often does not yet reflect the latest from today and yesterday.
- Crowdsourcing data (fresh view into the last 1-2 days): Risklayer GmbH crowdsource effort (see "Attribution" below):
- cases-rl-crowdsource-by-ags.csv and deaths-rl-crowdsource-by-ags.csv: per-Landkreis time series
- cases-rl-crowdsource-by-state.csv and deaths-rl-crowdsource-by-state.csv: per-Bundesland time series
- For the last ~48 hours these case count numbers (crowdsourced from Gesundheitsämter) may be a little more credible than what the RKI data set shows. For assessing the differences between the RKI data set(s) and the Risklayer data set(s) please also have a look at the plot above, and always try to do your own research.
- 7-day incidence time series resolved by county based on Risklayer data can be found in
more-data/.
- ags.json:
- for translating "amtlicher Gemeindeschlüssel" (AGS) to Landreis/Bundesland details, including latitude and longitude.
- containing per-county population data (see pull/383 for details).
- JSON endpoint /now: Germany's total case count (updated in real time, always fresh, for the sensationalists) -- Update Feb 2021: the HTTP API was disabled.
- data.csv: history, mixed data source based on RKI/ZEIT ONLINE. This did power the per-Bundesland time series exposed by the HTTP JSON API up until Jan 2021.
- It includes historical data for individual Bundesländer and Landkreise (states and counties).
- Its time series data is being re-written as data gets better over time. This is based on official RKI-provided time series data which receives daily updates even for days weeks in the past (accounting for delay in reporting).
Focus: predictable/robust machine readability. Backwards-compatibility (columns get added; but have never been removed so far).
- The column names use the ISO 3166 code for individual states.
- The points in time are encoded using localized ISO 8601 time string notation.
Note that the numbers for "today" as presented in media often actually refer to the last known state of data on the evening before. To address this ambiguity, the sample timestamps in the CSV files presented in this repository contain the time of the day (and not just the day). With that, consumers can have a vague impression about whether the sample represents the state in the morning or evening -- a common confusion / ambiguity with other data sets.
The recovered metric is not presented because it is rather blurry.
Feel free to consume it from other sources!
I tried to discover these step-by-step, they are possibly underrated (April 2020, minor updates towards the end of 2020):
- Bayern: case numbers, map, LK table
- Berlin: case numbers, map, intensive care numbers
- Baden-Württemberg:
- Brandenburg: press releases
- Bremen: press releases
- Hamburg: case numbers, press releases
- Hessen: press releases
- NRW: case numbers, LK table
- Mecklenburg-Vorpommern: press releases
- Niedersachsen (pretty well done!):
- case numbers, map, LK table
- CSV / GeoJSON
- so close, but no historical data :-(
- Rheinland-Pfalz: case numbers, LK table
- Saarland: case numbers
- Sachsen: case numbers, LK table, intensive care numbers
- Sachsen-Anhalt: case numbers, LK table, intensive care numbers
- Schleswig-Holstein: case numbers, LK table
- Thüringen: case numbers, LK table, intensive care numbers
- In this blog post (German) I try to shed light on why — as of the time of writing (March 18) — the numbers reported in the RKI and WHO situation reports lag behind by 1-3 days.
- Blog post Covid-19 HTTP API: German case numbers
- Blog post Covid-19 HTTP API: case numbers as time series, for individual German states
This is a very high-level changelog. Technical details of reporting changed all the time, most details can be inferred from GitHub issues.
- 2023-04: disabled daily updates
- 2022-05: the Risklayer crowdsourcing effort mentioned below has been discontinued in March 2022. Corresponding data files and plots are not updated anymore in this repository.
- 2021-02: I disabled the HTTP API. It's best to directly use the data files from this respository.
Please question the conclusiveness of these numbers. Some directions along which you may want to think:
- Germany seems to perform a large number of tests. But think about how much insight you actually have into how the testing rate (and its spatial distribution) evolves over time. In my opinion, one absolutely should know a whole lot about the testing effort itself before drawing conclusions from the time evolution of case count numbers.
- Each confirmed case is implicitly associated with a reporting date. We do not know for sure how that reporting date relates to the date of taking the sample.
- We believe that each "confirmed case" actually corresponds to a polymerase chain reaction (PCR) test for the SARS-CoV2 virus with a positive outcome. Well, I think that's true, we can have that much trust into the system.
- We seem to believe that the change of the number of confirmed COVID-19 cases over time is somewhat expressive: but what does it shed light on, exactly? The amount of testing performed, and its spatial coverage? The efficiency with which the virus spreads through the population ("basic reproduction number")? The actual, absolute number of people infected? The virus' potential to exhibit COVID-19 in an infected human body?
If you keep these (and more) ambiguities and questions in mind then I think you are ready to look at these numbers and their time evolution :-) 😷.
In Germany, every step along the chain of reporting (Meldekette) introduces a noticeable delay. This is not necessary, but sadly the current state of affairs. The Robert Koch-Institut (RKI) seems to be working on a more modern reporting system that might mitigate some of these delays along the Meldekette in the future. Until then, it is fair to assume that case numbers published by RKI have 1-2 days delay over the case numbers published by Landkreise, which themselves have an unknown lag relative to the physical tests. In some cases, the Meldekette might even be entirely disrupted, as discussed in this SPIEGEL article (German). Also see this discussion.
Wishlist: every case should be tracked with its own time line, and transparently change state over time. The individual cases (and their time lines) should be aggregated on a country-wide level, anonymously, and get published in almost real time, through an official, structured data source, free to consume for everyone.
Beginning of March 2020: shout-out to ZEIT ONLINE for continuously collecting and publishing the state-level data with little delay.
Edit March 21, 2020: Notably, by now the Berliner Morgenpost seems to do an equally well job of quickly aggregating the state-level data. We are using that in here, too. Thanks!
Edit March 26, 2020: Risklayer is coordinating a crowd-sourcing effort to process verified Landkreis data as quickly as possible. Tagesspiegel is verifying this effort and using it in their overview page. As far as I can tell this is so far the most transparent data flow, and also the fastest, getting us the freshest case count numbers. Great work!
Edit December 13, 2020: for the *-rl-crowdsource*.csv files proper legal attribution goes to
Risklayer GmbH (www.risklayer.com) and Center for Disaster Management and Risk Reduction Technology (CEDIM) at Karlsruhe Institute of Technology (KIT) and the Risklayer-CEDIM-Tagesspiegel SARS-CoV-2 Crowdsourcing Contributors