quantile-forest offers a Python implementation of quantile regression forests compatible with scikit-learn.
Quantile regression forests (QRF) are a non-parametric, tree-based ensemble method for estimating conditional quantiles, with application to high-dimensional data and uncertainty estimation [1]. The estimators in this package are performant, Cython-optimized QRF implementations that extend the forest estimators available in scikit-learn to estimate conditional quantiles. The estimators can estimate arbitrary quantiles at prediction time without retraining and provide methods for out-of-bag estimation, calculating quantile ranks, and computing proximity counts. They are compatible with and can serve as drop-in replacements for the scikit-learn variants.
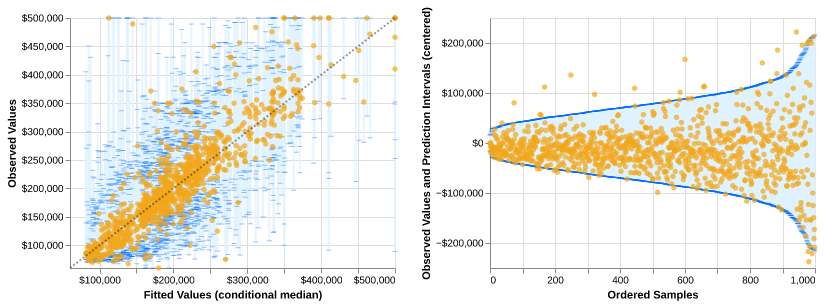
Example of fitted model predictions and prediction intervals on California housing data (code)
Install quantile-forest from PyPI using pip:
pip install quantile-forestfrom quantile_forest import RandomForestQuantileRegressor
from sklearn import datasets
X, y = datasets.fetch_california_housing(return_X_y=True)
qrf = RandomForestQuantileRegressor()
qrf.fit(X, y)
y_pred = qrf.predict(X, quantiles=[0.025, 0.5, 0.975])An installation guide, API documentation, and examples can be found in the documentation.
[1] N. Meinshausen, "Quantile Regression Forests", Journal of Machine Learning Research, 7(Jun), 983-999, 2006. http://www.jmlr.org/papers/volume7/meinshausen06a/meinshausen06a.pdf
If you use this package in academic work, please consider citing https://joss.theoj.org/papers/10.21105/joss.05976:
@article{Johnson2024,
doi = {10.21105/joss.05976},
url = {https://doi.org/10.21105/joss.05976},
year = {2024},
publisher = {The Open Journal},
volume = {9},
number = {93},
pages = {5976},
author = {Reid A. Johnson},
title = {quantile-forest: A Python Package for Quantile Regression Forests},
journal = {Journal of Open Source Software}
}