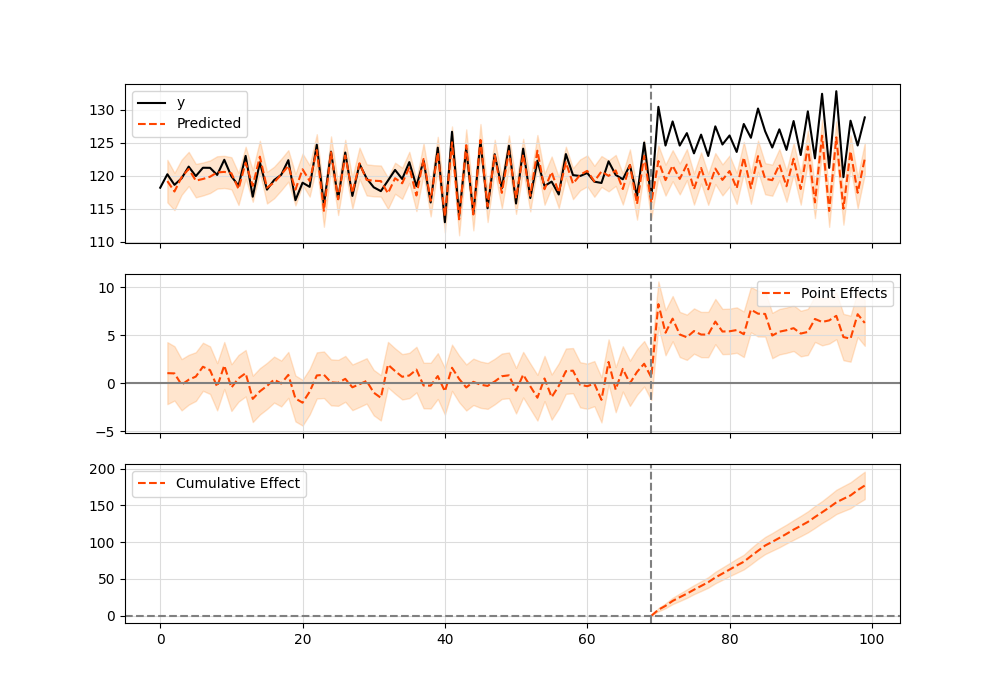
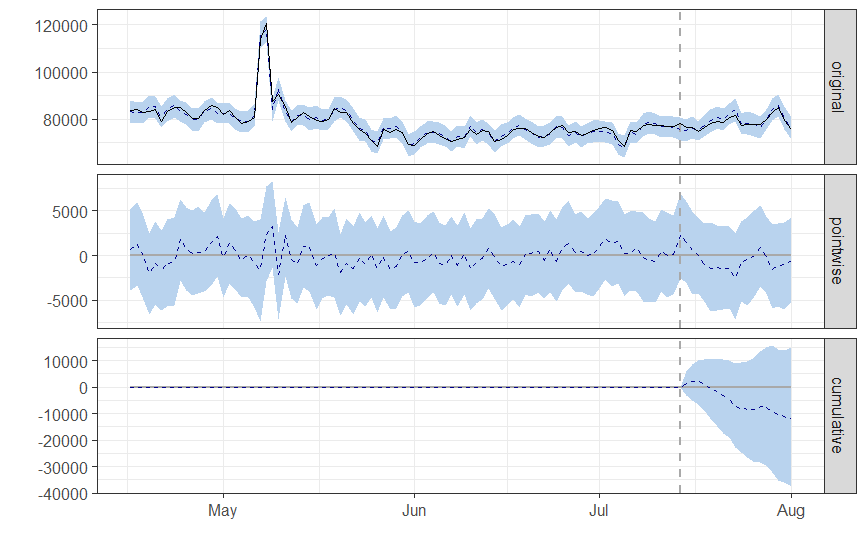
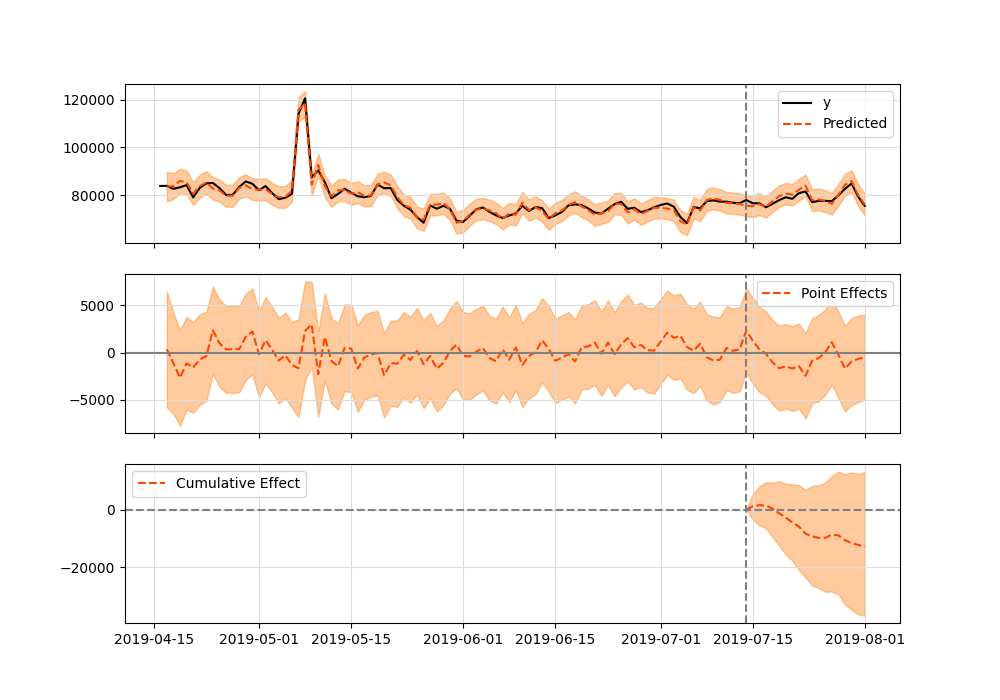
When I run the model, ci = CausalImpact(protests['count'], pre_period, post_period). This error happened
`---------------------------------------------------------------------------
NotImplementedError Traceback (most recent call last)
in
2 post_period = [73, len(protests)-1]
3
----> 4 ci = CausalImpact(protests['count'], pre_period, post_period)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/causalimpact/main.py in init(self, data, pre_period, post_period, model, model_args, alpha)
217 self.normed_post_data = processed_input['normed_post_data']
218 self.mu_sig = processed_input['mu_sig']
--> 219 self._fit_model()
220 self._process_posterior_inferences()
221 self._summarize_inferences()
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/causalimpact/main.py in _fit_model(self)
296 # if operation iloc returns a pd.Series, cast it back to pd.DataFrame
297 observed_time_series = pd.DataFrame(observed_time_series.iloc[:, 0])
--> 298 model_samples, model_kernel_results = cimodel.fit_model(
299 self.model,
300 observed_time_series,
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/causalimpact/model.py in fit_model(model, observed_time_series, method)
358 optimizer = tf.optimizers.Adam(learning_rate=0.1)
359 variational_steps = 200 # Hardcoded for now
--> 360 variational_posteriors = tfp.sts.build_factored_surrogate_posterior(model=model)
361
362 @tf.function()
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/sts/fitting.py in build_factored_surrogate_posterior(model, batch_shape, seed, name)
201 variational_posterior = collections.OrderedDict()
202 for param in model.parameters:
--> 203 variational_posterior[param.name] = _build_posterior_for_one_parameter(
204 param, batch_shape=batch_shape, seed=seed())
205 return joint_distribution_named_lib.JointDistributionNamed(
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/sts/fitting.py in _build_posterior_for_one_parameter(param, batch_shape, seed)
85
86 # Build a trainable Normal distribution.
---> 87 initial_loc = sample_uniform_initial_state(
88 param, init_sample_shape=batch_shape,
89 return_constrained=False, seed=seed)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/sts/fitting.py in sample_uniform_initial_state(parameter, return_constrained, init_sample_shape, seed)
67 parameter.prior.sample(init_sample_shape)))
68 param_shape = (
---> 69 unconstrained_prior_sample_fn.get_concrete_function().output_shapes)
70 if not tensorshape_util.is_fully_defined(param_shape):
71 param_shape = tf.shape(unconstrained_prior_sample_fn())
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/def_function.py in get_concrete_function(self, *args, **kwargs)
1297 ValueError: if this object has not yet been called on concrete values.
1298 """
-> 1299 concrete = self._get_concrete_function_garbage_collected(*args, **kwargs)
1300 concrete._garbage_collector.release() # pylint: disable=protected-access
1301 return concrete
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/def_function.py in _get_concrete_function_garbage_collected(self, *args, **kwargs)
1203 if self._stateful_fn is None:
1204 initializers = []
-> 1205 self._initialize(args, kwargs, add_initializers_to=initializers)
1206 self._initialize_uninitialized_variables(initializers)
1207
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/def_function.py in _initialize(self, args, kwds, add_initializers_to)
723 self._graph_deleter = FunctionDeleter(self._lifted_initializer_graph)
724 self._concrete_stateful_fn = (
--> 725 self._stateful_fn._get_concrete_function_internal_garbage_collected( # pylint: disable=protected-access
726 *args, **kwds))
727
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/function.py in _get_concrete_function_internal_garbage_collected(self, *args, **kwargs)
2967 args, kwargs = None, None
2968 with self._lock:
-> 2969 graph_function, _ = self._maybe_define_function(args, kwargs)
2970 return graph_function
2971
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/function.py in _maybe_define_function(self, args, kwargs)
3359
3360 self._function_cache.missed.add(call_context_key)
-> 3361 graph_function = self._create_graph_function(args, kwargs)
3362 self._function_cache.primary[cache_key] = graph_function
3363
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/function.py in _create_graph_function(self, args, kwargs, override_flat_arg_shapes)
3194 arg_names = base_arg_names + missing_arg_names
3195 graph_function = ConcreteFunction(
-> 3196 func_graph_module.func_graph_from_py_func(
3197 self._name,
3198 self._python_function,
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/framework/func_graph.py in func_graph_from_py_func(name, python_func, args, kwargs, signature, func_graph, autograph, autograph_options, add_control_dependencies, arg_names, op_return_value, collections, capture_by_value, override_flat_arg_shapes)
988 _, original_func = tf_decorator.unwrap(python_func)
989
--> 990 func_outputs = python_func(*func_args, **func_kwargs)
991
992 # invariant: func_outputs contains only Tensors, CompositeTensors,
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/def_function.py in wrapped_fn(*args, **kwds)
632 xla_context.Exit()
633 else:
--> 634 out = weak_wrapped_fn().wrapped(*args, **kwds)
635 return out
636
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/framework/func_graph.py in wrapper(*args, **kwargs)
975 except Exception as e: # pylint:disable=broad-except
976 if hasattr(e, "ag_error_metadata"):
--> 977 raise e.ag_error_metadata.to_exception(e)
978 else:
979 raise
NotImplementedError: in user code:
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/sts/fitting.py:66 None *
parameter.prior.sample(init_sample_shape))
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/distributions/distribution.py:1002 sample **
return self._call_sample_n(sample_shape, seed, name, **kwargs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/distributions/transformed_distribution.py:331 _call_sample_n
x = self.distribution.sample(sample_shape=[n], seed=seed,
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/distributions/distribution.py:1002 sample
return self._call_sample_n(sample_shape, seed, name, **kwargs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/distributions/distribution.py:979 _call_sample_n
samples = self._sample_n(
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/internal/distribution_util.py:1364 _fn
return fn(*args, **kwargs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/distributions/inverse_gamma.py:209 _sample_n
return tf.math.exp(-gamma_lib.random_gamma(
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/distributions/gamma.py:660 random_gamma
return random_gamma_with_runtime(
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/distributions/gamma.py:654 random_gamma_with_runtime
return _random_gamma_gradient(
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/internal/custom_gradient.py:104 none_wrapper
return f_wrapped(*trimmed_args, **kwargs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/ops/custom_gradient.py:261 __call__
return self._d(self._f, a, k)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/ops/custom_gradient.py:217 decorated
return _graph_mode_decorator(wrapped, args, kwargs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/ops/custom_gradient.py:330 _graph_mode_decorator
result, grad_fn = f(*args)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/internal/custom_gradient.py:92 f_wrapped
val, aux = vjp_fwd(*reconstruct_args, **kwargs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/distributions/gamma.py:541 _random_gamma_fwd
samples, impl = _random_gamma_no_gradient(
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/internal/implementation_selection.py:83 f_wrapped
return f(*args, **kwargs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/def_function.py:828 __call__
result = self._call(*args, **kwds)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/def_function.py:862 _call
results = self._stateful_fn(*args, **kwds)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/function.py:2941 __call__
filtered_flat_args) = self._maybe_define_function(args, kwargs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/function.py:3361 _maybe_define_function
graph_function = self._create_graph_function(args, kwargs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/function.py:3196 _create_graph_function
func_graph_module.func_graph_from_py_func(
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/framework/func_graph.py:990 func_graph_from_py_func
func_outputs = python_func(*func_args, **func_kwargs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/def_function.py:634 wrapped_fn
out = weak_wrapped_fn().__wrapped__(*args, **kwds)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/distributions/gamma.py:483 _random_gamma_no_gradient
return sampler_impl(
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow_probability/python/internal/implementation_selection.py:162 impl_selecting_fn
function.register(defun_cpu_fn, **kwargs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/function.py:3390 register
concrete_func.add_gradient_functions_to_graph()
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/function.py:2057 add_gradient_functions_to_graph
self._delayed_rewrite_functions.forward_backward())
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/function.py:631 forward_backward
forward, backward = self._construct_forward_backward(num_doutputs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/function.py:674 _construct_forward_backward
func_graph_module.func_graph_from_py_func(
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/framework/func_graph.py:990 func_graph_from_py_func
func_outputs = python_func(*func_args, **func_kwargs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/eager/function.py:665 _backprop_function
return gradients_util._GradientsHelper( # pylint: disable=protected-access
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/ops/gradients_util.py:683 _GradientsHelper
in_grads = _MaybeCompile(grad_scope, op, func_call,
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/ops/gradients_util.py:340 _MaybeCompile
return grad_fn() # Exit early
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/ops/gradients_util.py:684 <lambda>
lambda: grad_fn(op, *out_grads))
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/ops/random_grad.py:114 _StatelessRandomGammaV2Grad
alpha_broadcastable = add_leading_unit_dimensions(alpha,
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/ops/random_grad.py:35 add_leading_unit_dimensions
[array_ops.ones([num_dimensions], dtype=dtypes.int32),
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/util/dispatch.py:201 wrapper
return target(*args, **kwargs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/ops/array_ops.py:3120 ones
output = _constant_if_small(one, shape, dtype, name)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/ops/array_ops.py:2804 _constant_if_small
if np.prod(shape) < 1000:
<__array_function__ internals>:5 prod
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/numpy/core/fromnumeric.py:3030 prod
Returns
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/numpy/core/fromnumeric.py:87 _wrapreduction
return ufunc.reduce(obj, axis, dtype, out, **passkwargs)
/Library/Frameworks/Python.framework/Versions/3.8/lib/python3.8/site-packages/tensorflow/python/framework/ops.py:852 __array__
raise NotImplementedError(
NotImplementedError: Cannot convert a symbolic Tensor (gradients/stateless_random_gamma/StatelessRandomGammaV2_grad/sub:0) to a numpy array. This error may indicate that you're trying to pass a Tensor to a NumPy call, which is not supported`