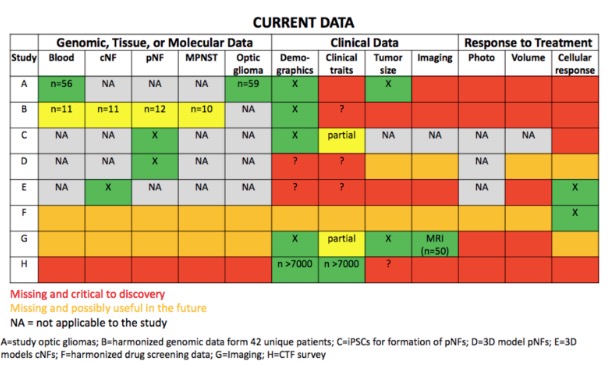
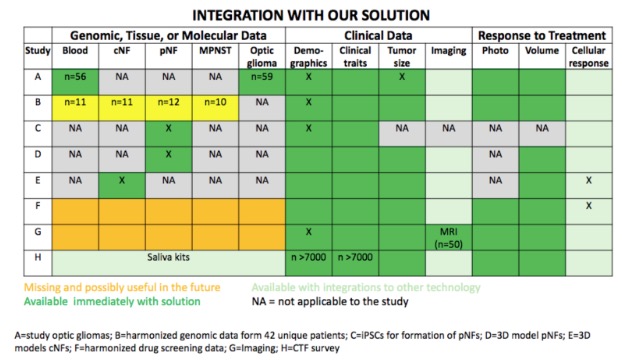
Dermaviz: Comprehensive Research Suite for integrated Phenotyping, Genotyping and 3D Modeling for NF patients
Dermaviz is a full research suite designed to advance and integrate the collection of data from NF patients.
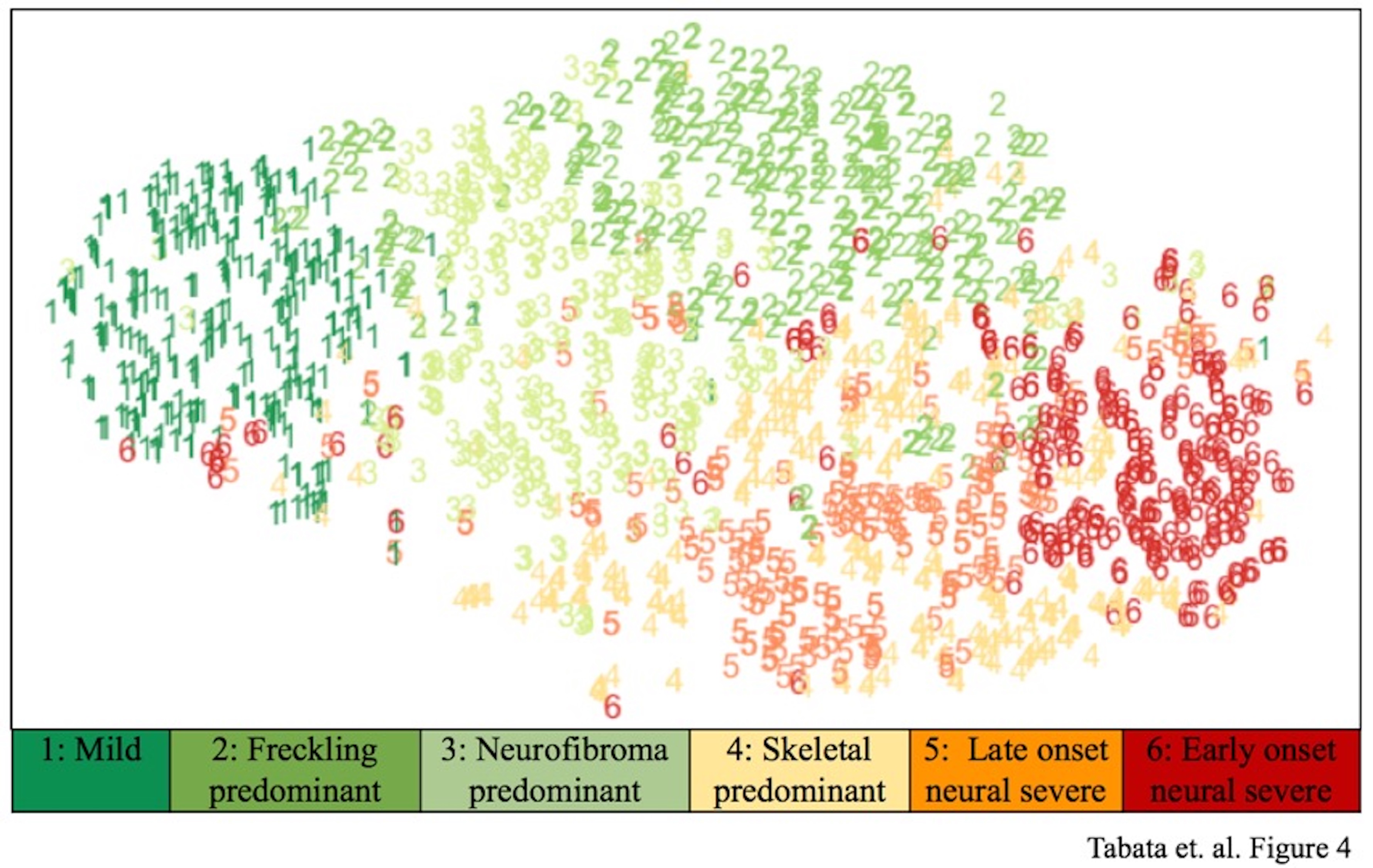
Neurofibromatosis type 1 (NF1) displays considerable variability in phenotypic expression and recent work identified phenotypic subtypes of disease. Large scale phenotypic-genotypic correlations will lead to better understanding of disease subtypes and more personalized screening and treatment of NF1. We have created an easy-to-use iOS mobile application that creates and exports a 3D model to HIPAA compliant cloud storage, integrates phenotypic and genomic data, and enables fast and comprehensive data collection for a large number of NF patients (>7,000). This solution will enable rapid collection of integrated patient data, leading to understanding of disease, identification of genetic modifiers, improved risk stratification, and measurement of response to therapeutics.
- The heterogeneity of NF1 is not understood
- Rich NF1 datasets are not linked to comprehensive associated phenotypic data
- Large sample size of linked genotype-phenotype data needed to understand heterogeneity and subtypes of disease
Neurofibromatosis (NF) type 1 is a highly heterogeneous disease. Currently, we do not have ways to predict which patients will develop certain characteristics or respond to therapies. In order to improve management of NF1, including both risk stratification and targeted therapeutics, a better understanding of disease heterogeneity is needed. A recent study by Tabata et. al. (under review) identified phenotypic subtypes. However, genotypic and phenotypic data is needed from a large sample size (>2000) in order to perform genome-wide association studies to identify novel modifiers of disease to further understand heterogeneity. Our work creates a novel method of data collection that will enable rapid data collection of phenotypic, genotypic, and 3D patient data in order to rapidly advance research for NF1. This solution will enable personalized risk stratification, development of therapeutics, and response to therapeutics. Integration of our solution with existing data will synergistically result in a powerful database for discovery. Furthermore, our solution is scalable, HIPAA compliant, and translatable to other rare diseases.
-
Mobile application generates high resolution 3D model of NF1 patient’s body
-
Rapidly collect thousands of 3D images of NF1 patients’ skin
-
Store in HIPAA-compliant Google Cloud folder for phenotypic visualization and downstream ML
-
Genotype-phenotype research pipeline
A comprehensive patient survey has been adapted from the Children’s Tumor Foundation and formatted as a google forms survey. Patients will submit this form through the app, which will be linked to the patient’s unique cloud file. Genomic data is also on this cloud file.
After completing the patient data survey, the patient is assigned a unique invite code. The patient provides this in our app to submit their 3D scan. The app guides the patient on scanning their face and assembles into a 3D model. This model is sent up to the HIPAA-compliant server securely using HTTPS and is available in multiple popular file formats. These scans are sorted by date.
Google Cloud Servers are used for storage of patient phenotypic, genotypic, and 3D data. This can be accessed by dermatologists and researchers with access permissions and can be linked to patient electronic medical record data, such as MRI and lab results.
All genomic data and 3D models are stored by date and are available in a single patient file. Files are accessible by authenticated clinicians.
A pipeline is available for associating this data to Google BigQuery as well as assembling a dataset and sending over to any Machine Learning ready stack.
We created an iOS app that takes a high-resolution 3D scan of a patient’s face and exports it to a HIPAA-compliant Google Cloud server. We also developed a pipeline for connecting all collected 3D models to Google BigQuery as well to Machine-Learning ready stacks.
3D scan of team members’ faces successfully captured the makeshift “neurofibromas” that were placed on the skin. Furthermore, the 3D face image was successfully exported to the Google Cloud server and linked to clinical, genomic, and survey data.
Our suite integrates with a patient questionnaire and genomic data collection. Each patient is assigned a unique invite code that allows them to “log-in” to the app and take scans. All scans are securely uploaded to our HIPAA-compliant Google Cloud server over HTTPS. Each scan is sorted by date taken to provide clinicians with a comprehensive view on the development of NF over time. These scans are available in the standard .obj format. The dermatologist will also have access to patient data that includes submitted answers in the questionnaire as well the genomic data collected.
Once the form data and genomic data is available, the Dermaviz server generates an invite code and sends it to the patient. On the app, the patient enters the invite code. This code is cached client side and is used to send all data to the server.
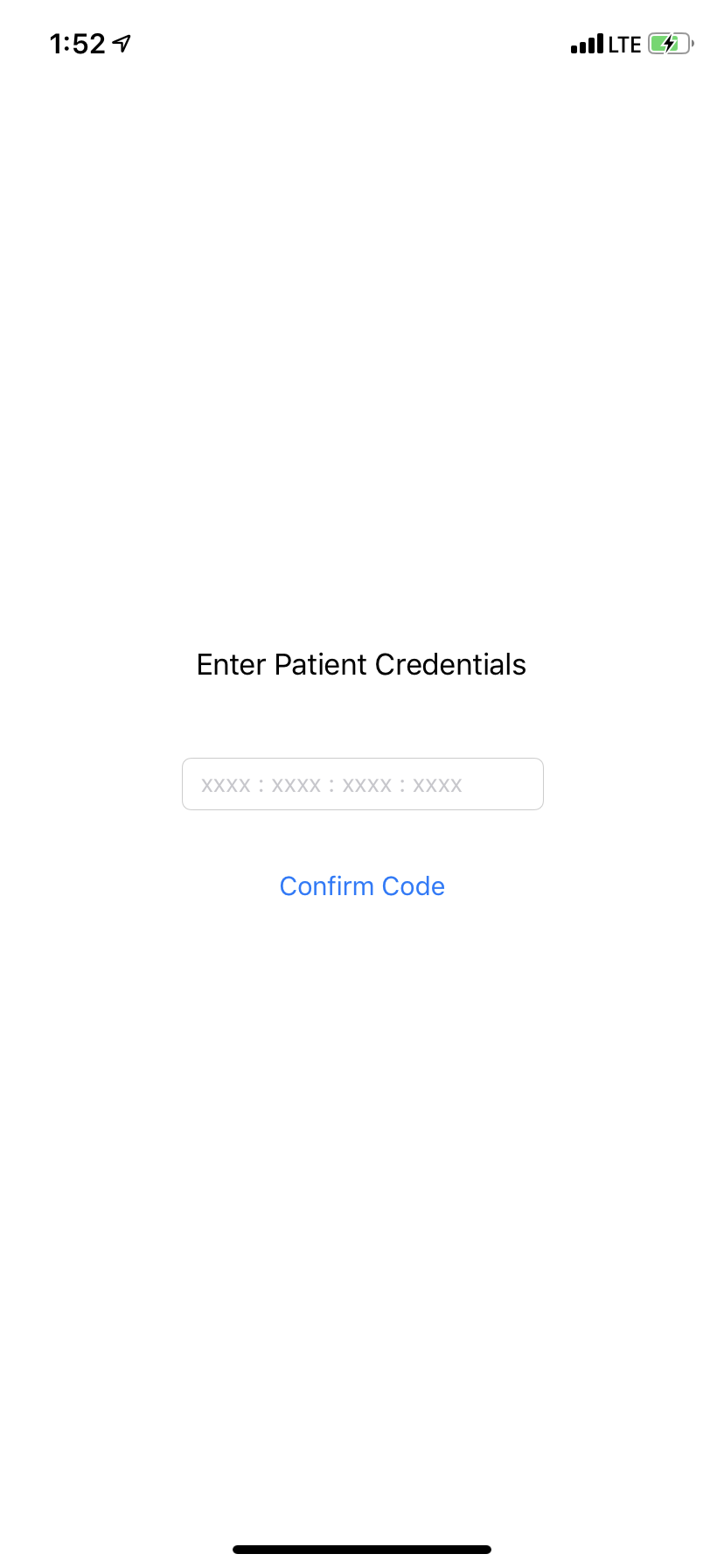
After entering the invite code, patients are instructed to scan their face. After a successful scan, these scans are uploaded to our server.
A phone application that captures high-resolution 3D images of NF1 patient skin allows for rapid collection of phenotypic NF1 data. We created here an iOS app that takes a high resolution 3D scan of the patient’s face, links it to patient clinical, genomic, and survey data, and exports it to a HIPAA-compliant Google Cloud folder for visualization by dermatologists and NF1 researchers. Rapid collection of phenotypic data and linking with clinical, genomic, and survey data enables robust characterizations of disease heterogeneity and will dramatically accelerate research in NF1 in the following areas:
-
Increase utility of currently available datasets
-
Obtaining prognostic information on who might need treatment
-
Capturing the natural history of NF1
-
Measuring cutaneous tumor burden over time
-
Tracking response to future therapies
-
Identifying pathways that modulate number of neurofibromas, which will present new genetic and molecular targets for drug therapy
-
Extrapolate to any rare cutaneous disease (Gorlin’s, dysplastic nevus syndrome, phakomatosis pigmentokeratotica, tuberous sclerosis, RAS-opathies, etc.)
Additional data that we will acquire with our solution includes phenotypic data, collected through the “Patient questionnaire of comprehensive NF1 clinical traits” (Questionnaire).
Whole genome sequencing data will be obtained through saliva samples, collected by mailing saliva kits to NF1 patients in collaboration with the CTF. We also plan to implement dermatologist portal for access to 3D models.
Measurement of responses to future therapies will be enabled by clinician or researcher input of drug information and future models into the patient’s file. Additionally, increased utilization of the app will allow us to gather image training data for machine learning advances off of 3D models for automated lesion counting.
- Expand to the rest of the body
- Human in the loop characterization to label training data for automated lesion count, volume burden, and classification
- Implement dermatologist portal for access to 3D models
- Implement a drag-and-drop model system to connect models created by researchers to clinical data
Our most critical next step is expansion to the rest of the body. We plan to automate lesion count, volume burden calculation, and classification of lesions by using human-in-the-loop characterization to label training data. With increased utilization of the app, we will have datasets with which to develop machine learning/ deep learning pipelines for image training to eventually fully automate lesion count, volume burden calculation, and classification of lesions. These deep learning pipelines will also improve reconstruction of 3D images for the rest of the body. Tools and skills needed to accomplish these future steps include: deep learning experience to enable training and reconstruction of high-resolution images for the rest of the body, experience with machine learning to implement human in the loop characterization, and iOS app development experience to implement new features.
Our current solution, including collection of patient saliva samples and clinical characteristics, will increase the utility of already-available data by creating a centralized space to store corresponding clinical with molecular and genetic data. Additional implementation of the future steps we described above will revolutionize data collection and allow for rapid research advancements for NF1.
We plan to release this app on the App Store and connect with labs and hospitals to create patient solutions.
| Name | Role |
|---|---|
| Mika Tabata (Stanford, Sarin Lab) | NF Researcher |
| Warren Ho Chan (Stanford, Sarin Lab) | NF Researcher |
| Rushil Srivastava (Stanford, Sarin Lab) | App Development & Backend Development |
| Michael Yan (Stanford) | App Development |
| Nam Nguyen (Stanford) | NF Researcher |
| Neekon Saadat (Stanford, Snyder Lab) | App Development |
Thanks to Dr. Kavita Sarin for her mentorship and guidance.
Special thanks to Bellus3D for licensing their 3D technology for us to research and build upon.
Logo credits to RoundIcons.