molecularai / chemformer Goto Github PK
View Code? Open in Web Editor NEWLicense: Apache License 2.0
License: Apache License 2.0
I have trouble reproducing the results of the paper, and I wanted to know if there was anything that I was doing wrong. I evaluated all the fine-tuned models provided (https://az.app.box.com/s/7eci3nd9vy0xplqniitpk02rbg9q2zcq/folder/145316934583) on the USPTO-50k dataset, and the highest accuracy I can report was using the USPTO-SEP model.
Could you please elaborate on how the results were obtained and possibly share the list of reactants it was tested on? I could have somehow chosen a problematic test split that led to poor results.
Apologies if that's a dumb question, I wanted to look into the models.py file from readme but wasn't able to find it in this repository. Could you, please, help me with this? The readme says the following: "Models: The models.py file contains a Pytorch Lightning implementation of the BART language model, as well as Pytorch Lightning implementations of models for downstream tasks.", but I couldn't locate the models.py file.
Hi!
In the experment of desirable molecules generation, Chemformer used beam search to generateoutput molecules, while the Transformer and Transformer-R used greedy search. Why make sampling different? Or each sampling method will produce same result?
Is this model can make SMILES to vector?
How to do it?
best regard
I was running into several issues with the described setup.
First cuda 11.1 did not work with the described fairscale. I resolved that by using 0.4.1. I also dropped optuna for now because the installation seems to downgrade pytorch. Just wanted to leave it here in case others run into similar issues. It seems to run now.
Hi there, thanks for your great work!
I found that in predict.py you directly use the input smiles string rather than a canonical one for inference. Is this intended to be that? If so, will two smiles string for the same molecule lead to different predictions?
Best regards!
Hi,
I failed to install Chemformer as module, and it seems to be because setup.py is missing packages=find_packages(exclude=[]). Could you maybe add that? It would ease our setup considerably. Thank you so much already!
I've been working on using the encoder of a pre-trained transformer model to extract the 2D memory tensor representation of a SMILES molecule. After some initial challenges, I've successfully configured the model to generate these memory tensors.
It appears that the encoder expects an input vector where each element represents the IDs of the corresponding token in the SMILES sequence. To accommodate variations in SMILES length, the input vector is padded to a fixed length of 512 using the ID of the padding character (i.e., 0).
However, I'm still unsure about the specific role of the padding mask vector. Does it indicate true values at padded positions or where the actual SMILES sequence resides?
I have been unable to find some of the datasets that appear to be essential for the running of the code, such as the "uspto_50.txt" file. I converted the uspto_50.pickle to text in a tabulated format and changed the "reactants_mol" and "products_mol" columns to "reactants" and "products", respectively, but then the program could not find a sequence length column: KeyError: 'This dataset does not store any sequence lengths'.
Similarly, while while running predict.sh, I couldn't find the "bart_vocab_downstream.txt" file, only "bart_vocab_downstream.json file is available.
Could you please point me to the correct files: uspto_50.txt and bart_vocab_downstream.txt?
Hello,
I would like to get the embeddings (output from BART encoder) and separately use the embeddings as an input to the decoder ti get back SMILES string as presented in step 1 in Figure1 in the paper. I don't see a direct way, could you please suggest me any methods?
Hi All,
Hope that you are okay. Just to provide a bit of background, I have been trying to run the finetuneRegr.py script in the conda command line in order to fine-tune the model and predict the conductivity of some compounds. I formatted my data file according to the steps detailed in #13 and specified the paths to my 1.data, 2. one of the models found in the link, and 3. the vocabulary text file with the added token.
After jumping through some hurdles (mostly changing some lines to account for the updated packages), I am now stuck after I get an error saying that the program needs [0] GPUs to run and I have: []. Could you please confirm that I'm following the right procedure for the task at hand? Are you aware of any way to get around this issue (such as changing the code or using a Cloud GPU Service?
Kind regards,
Diego
while running molbart.predict in macos I got the error i received the error AttributeError: module 'os' has no attribute 'sched_getaffinity' in molbart/modules/data/base.py. Similary I received error `AttributeError: module 'os' has no attribute 'sched_getaffinity' in molbart/models/chemformer.py. Currently I handled the issue with
try:
self._num_workers = len(os.sched_getaffinity(0))
except AttributeError:
self._num_workers = os.cpu_count() or 1
try:
n_cpus = len(os.sched_getaffinity(0))
except AttributeError:
n_cpus = os.cpu_count() or 1 # Default to 1 if cpu_count() is None
I am wondering if this serves the original purpose of sched_getaffinity(0) and is there a better way to handle it in mac ?
Hello,
When attempting to fine-tune in a multi-gpu SLURM environment, I get the following error:
Mon Sep 18 09:20:06 2023
+---------------------------------------------------------------------------------------+
| NVIDIA-SMI 535.104.05 Driver Version: 535.104.05 CUDA Version: 12.2 |
|-----------------------------------------+----------------------+----------------------+
| GPU Name Persistence-M | Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap | Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|=========================================+======================+======================|
| 0 Tesla V100-SXM2-32GB On | 00000000:1A:00.0 Off | 0 |
| N/A 31C P0 44W / 300W | 0MiB / 32768MiB | 0% E. Process |
| | | N/A |
+-----------------------------------------+----------------------+----------------------+
| 1 Tesla V100-SXM2-32GB On | 00000000:1C:00.0 Off | 0 |
| N/A 27C P0 41W / 300W | 0MiB / 32768MiB | 0% E. Process |
| | | N/A |
+-----------------------------------------+----------------------+----------------------+
| 2 Tesla V100-SXM2-32GB On | 00000000:1D:00.0 Off | 0 |
| N/A 28C P0 42W / 300W | 0MiB / 32768MiB | 0% E. Process |
| | | N/A |
+-----------------------------------------+----------------------+----------------------+
| 3 Tesla V100-SXM2-32GB On | 00000000:1E:00.0 Off | 0 |
| N/A 30C P0 42W / 300W | 0MiB / 32768MiB | 0% E. Process |
| | | N/A |
+-----------------------------------------+----------------------+----------------------+
+---------------------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=======================================================================================|
| No running processes found |
+---------------------------------------------------------------------------------------+
MASTER_ADDR=mg089
GpuFreq=control_disabled
INFO: underlay of /etc/localtime required more than 50 (80) bind mounts
INFO: underlay of /etc/localtime required more than 50 (80) bind mounts
INFO: underlay of /etc/localtime required more than 50 (80) bind mounts
INFO: underlay of /etc/localtime required more than 50 (80) bind mounts
INFO: underlay of /usr/bin/nvidia-smi required more than 50 (251) bind mounts
INFO: underlay of /usr/bin/nvidia-smi required more than 50 (251) bind mounts
INFO: underlay of /usr/bin/nvidia-smi required more than 50 (251) bind mounts
INFO: underlay of /usr/bin/nvidia-smi required more than 50 (251) bind mounts
GPU available: True, used: True
TPU available: None, using: 0 TPU cores
Multi-processing is handled by Slurm.
GPU available: True, used: True
TPU available: None, using: 0 TPU cores
Multi-processing is handled by Slurm.
GPU available: True, used: True
TPU available: None, using: 0 TPU cores
Multi-processing is handled by Slurm.
GPU available: True, used: True
TPU available: None, using: 0 TPU cores
Multi-processing is handled by Slurm.
/opt/conda/lib/python3.8/site-packages/pytorch_lightning/utilities/distributed.py:50: UserWarning: you defined a validation_step but have no val_dataloader. Skipping validation loop
warnings.warn(*args, **kwargs)
initializing ddp: GLOBAL_RANK: 1, MEMBER: 2/4
/opt/conda/lib/python3.8/site-packages/pytorch_lightning/utilities/distributed.py:50: UserWarning: you defined a validation_step but have no val_dataloader. Skipping validation loop
warnings.warn(*args, **kwargs)
initializing ddp: GLOBAL_RANK: 3, MEMBER: 4/4
/opt/conda/lib/python3.8/site-packages/pytorch_lightning/utilities/distributed.py:50: UserWarning: you defined a validation_step but have no val_dataloader. Skipping validation loop
warnings.warn(*args, **kwargs)
initializing ddp: GLOBAL_RANK: 2, MEMBER: 3/4
/opt/conda/lib/python3.8/site-packages/pytorch_lightning/utilities/distributed.py:50: UserWarning: you defined a validation_step but have no val_dataloader. Skipping validation loop
warnings.warn(*args, **kwargs)
initializing ddp: GLOBAL_RANK: 0, MEMBER: 1/4
You have not specified an optimizer or scheduler within the DeepSpeed config.Using `configure_optimizers` to define optimizer and scheduler.
INFO:lightning:You have not specified an optimizer or scheduler within the DeepSpeed config.Using `configure_optimizers` to define optimizer and scheduler.
Traceback (most recent call last):
File "/opt/conda/lib/python3.8/runpy.py", line 194, in _run_module_as_main
return _run_code(code, main_globals, None,
File "/opt/conda/lib/python3.8/runpy.py", line 87, in _run_code
exec(code, run_globals)
File "/ui/abv/hoangsx/Chemformer/temp/train_boring.py", line 77, in <module>
run()
File "/ui/abv/hoangsx/Chemformer/temp/train_boring.py", line 72, in run
trainer.fit(model, train_data)
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py", line 511, in fit
self.pre_dispatch()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py", line 540, in pre_dispatch
self.accelerator.pre_dispatch()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/accelerators/accelerator.py", line 84, in pre_dispatch
self.training_type_plugin.pre_dispatch()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/plugins/training_type/deepspeed.py", line 174, in pre_dispatch
self.init_deepspeed()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/plugins/training_type/deepspeed.py", line 193, in init_deepspeed
self._initialize_deepspeed_train(model)
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/plugins/training_type/deepspeed.py", line 218, in _initialize_deepspeed_train
model, optimizer, _, lr_scheduler = deepspeed.initialize(
File "/opt/conda/lib/python3.8/site-packages/deepspeed/__init__.py", line 112, in initialize
engine = DeepSpeedEngine(args=args,
File "/opt/conda/lib/python3.8/site-packages/deepspeed/runtime/engine.py", line 145, in __init__
self._configure_with_arguments(args, mpu)
File "/opt/conda/lib/python3.8/site-packages/deepspeed/runtime/engine.py", line 499, in _configure_with_arguments
args.local_rank = int(os.environ['LOCAL_RANK'])
File "/opt/conda/lib/python3.8/os.py", line 675, in __getitem__
raise KeyError(key) from None
KeyError: 'LOCAL_RANK'
N GPUS: 4
N NODES: 1
You have not specified an optimizer or scheduler within the DeepSpeed config.Using `configure_optimizers` to define optimizer and scheduler.
INFO:lightning:You have not specified an optimizer or scheduler within the DeepSpeed config.Using `configure_optimizers` to define optimizer and scheduler.
You have not specified an optimizer or scheduler within the DeepSpeed config.Using `configure_optimizers` to define optimizer and scheduler.
INFO:lightning:You have not specified an optimizer or scheduler within the DeepSpeed config.Using `configure_optimizers` to define optimizer and scheduler.
You have not specified an optimizer or scheduler within the DeepSpeed config.Using `configure_optimizers` to define optimizer and scheduler.
INFO:lightning:You have not specified an optimizer or scheduler within the DeepSpeed config.Using `configure_optimizers` to define optimizer and scheduler.
N GPUS: 4
N NODES: 1
Traceback (most recent call last):
File "/opt/conda/lib/python3.8/runpy.py", line 194, in _run_module_as_main
return _run_code(code, main_globals, None,
File "/opt/conda/lib/python3.8/runpy.py", line 87, in _run_code
exec(code, run_globals)
File "/ui/abv/hoangsx/Chemformer/temp/train_boring.py", line 77, in <module>
run()
File "/ui/abv/hoangsx/Chemformer/temp/train_boring.py", line 72, in run
trainer.fit(model, train_data)
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py", line 511, in fit
self.pre_dispatch()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py", line 540, in pre_dispatch
self.accelerator.pre_dispatch()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/accelerators/accelerator.py", line 84, in pre_dispatch
self.training_type_plugin.pre_dispatch()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/plugins/training_type/deepspeed.py", line 174, in pre_dispatch
self.init_deepspeed()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/plugins/training_type/deepspeed.py", line 193, in init_deepspeed
self._initialize_deepspeed_train(model)
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/plugins/training_type/deepspeed.py", line 218, in _initialize_deepspeed_train
model, optimizer, _, lr_scheduler = deepspeed.initialize(
File "/opt/conda/lib/python3.8/site-packages/deepspeed/__init__.py", line 112, in initialize
engine = DeepSpeedEngine(args=args,
File "/opt/conda/lib/python3.8/site-packages/deepspeed/runtime/engine.py", line 145, in __init__
self._configure_with_arguments(args, mpu)
File "/opt/conda/lib/python3.8/site-packages/deepspeed/runtime/engine.py", line 499, in _configure_with_arguments
args.local_rank = int(os.environ['LOCAL_RANK'])
File "/opt/conda/lib/python3.8/os.py", line 675, in __getitem__
raise KeyError(key) from None
KeyError: 'LOCAL_RANK'
N GPUS: 4
N NODES: 1
Traceback (most recent call last):
File "/opt/conda/lib/python3.8/runpy.py", line 194, in _run_module_as_main
return _run_code(code, main_globals, None,
File "/opt/conda/lib/python3.8/runpy.py", line 87, in _run_code
exec(code, run_globals)
File "/ui/abv/hoangsx/Chemformer/temp/train_boring.py", line 77, in <module>
run()
File "/ui/abv/hoangsx/Chemformer/temp/train_boring.py", line 72, in run
trainer.fit(model, train_data)
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py", line 511, in fit
self.pre_dispatch()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py", line 540, in pre_dispatch
self.accelerator.pre_dispatch()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/accelerators/accelerator.py", line 84, in pre_dispatch
self.training_type_plugin.pre_dispatch()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/plugins/training_type/deepspeed.py", line 174, in pre_dispatch
self.init_deepspeed()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/plugins/training_type/deepspeed.py", line 193, in init_deepspeed
self._initialize_deepspeed_train(model)
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/plugins/training_type/deepspeed.py", line 218, in _initialize_deepspeed_train
model, optimizer, _, lr_scheduler = deepspeed.initialize(
File "/opt/conda/lib/python3.8/site-packages/deepspeed/__init__.py", line 112, in initialize
engine = DeepSpeedEngine(args=args,
File "/opt/conda/lib/python3.8/site-packages/deepspeed/runtime/engine.py", line 145, in __init__
self._configure_with_arguments(args, mpu)
File "/opt/conda/lib/python3.8/site-packages/deepspeed/runtime/engine.py", line 499, in _configure_with_arguments
args.local_rank = int(os.environ['LOCAL_RANK'])
File "/opt/conda/lib/python3.8/os.py", line 675, in __getitem__
raise KeyError(key) from None
KeyError: 'LOCAL_RANK'
N GPUS: 4
N NODES: 1
Traceback (most recent call last):
File "/opt/conda/lib/python3.8/runpy.py", line 194, in _run_module_as_main
return _run_code(code, main_globals, None,
File "/opt/conda/lib/python3.8/runpy.py", line 87, in _run_code
exec(code, run_globals)
File "/ui/abv/hoangsx/Chemformer/temp/train_boring.py", line 77, in <module>
run()
File "/ui/abv/hoangsx/Chemformer/temp/train_boring.py", line 72, in run
trainer.fit(model, train_data)
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py", line 511, in fit
self.pre_dispatch()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/trainer/trainer.py", line 540, in pre_dispatch
self.accelerator.pre_dispatch()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/accelerators/accelerator.py", line 84, in pre_dispatch
self.training_type_plugin.pre_dispatch()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/plugins/training_type/deepspeed.py", line 174, in pre_dispatch
self.init_deepspeed()
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/plugins/training_type/deepspeed.py", line 193, in init_deepspeed
self._initialize_deepspeed_train(model)
File "/opt/conda/lib/python3.8/site-packages/pytorch_lightning/plugins/training_type/deepspeed.py", line 218, in _initialize_deepspeed_train
model, optimizer, _, lr_scheduler = deepspeed.initialize(
File "/opt/conda/lib/python3.8/site-packages/deepspeed/__init__.py", line 112, in initialize
engine = DeepSpeedEngine(args=args,
File "/opt/conda/lib/python3.8/site-packages/deepspeed/runtime/engine.py", line 145, in __init__
self._configure_with_arguments(args, mpu)
File "/opt/conda/lib/python3.8/site-packages/deepspeed/runtime/engine.py", line 499, in _configure_with_arguments
args.local_rank = int(os.environ['LOCAL_RANK'])
File "/opt/conda/lib/python3.8/os.py", line 675, in __getitem__
raise KeyError(key) from None
KeyError: 'LOCAL_RANK'
My SLURM job request is as follows:
#!/bin/bash
#SBATCH --job-name=chemformer
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=4
#SBATCH -p gpu
#SBATCH --gres=gpu:4
#SBATCH --mem=64gb
#SBATCH --time=0:15:00
#SBATCH --output=slurm_out/output.%A_%a.txt
module load miniconda3
module load cuda/11.8.0
nvidia-smi
export MASTER_PORT=$(expr 10000 + $(echo -n $SLURM_JOBID | tail -c 4))
export WORLD_SIZE=$(($SLURM_NNODES * $SLURM_NTASKS_PER_NODE))
echo "WORLD_SIZE="$WORLD_SIZE
master_addr=$(scontrol show hostnames "$SLURM_JOB_NODELIST" | head -n 1)
export MASTER_ADDR=$master_addr
echo "MASTER_ADDR="$MASTER_ADDR
CONTAINER="singularity exec --nv $PROJ_DIR/singularity_images/chemformer.sif"
CMD="python -m molbart.fine_tune \
--dataset uspto_50 \
--data_path data/seq-to-seq_datasets/uspto_50.pickle \
--model_path models/pre-trained/combined/step=1000000.ckpt \
--task backward_prediction \
--epochs 100 \
--lr 0.001 \
--schedule cycle \
--batch_size 128 \
--acc_batches 4 \
--augment all \
--aug_prob 0.5 \
--gpus $SLURM_NTASKS_PER_NODE
"
srun ${CONTAINER} ${CMD}
I am using the original library versions defined in the documentation. Is this behavior expected?
Hello, I've been looking to run predictions through your fine-tuned USPTO-50K model, and I'm getting 0% accuracy. Even though the generated molecules are similar, not a single one is exactly correct. Here are the test SMILES I gave the model as input. Here's an example of the output I'm getting:
python -m molbart.predict \
--reactants_path {input_file} \
--products_path {output_file} \
--model_path {model_file} \
--batch_size 64 \
--n_beams 10source | COC(=O)c1ccc2c(c1)n(CCC(C)C)c(=O)n2CCC(C)C
prediction | CC(C)CCBr.COC(=O)c1ccc2[nH]c(=O)n(CCC(C)C)c2c1
target | CC(C)CCn1c(=O)n(CCC(C)C)c2cc(C(=O)O)ccc21
The sha1sum of the checkpoint i downloaded is c859c68b198ac1b8cfab48196bdde6b35641bf81. However, I had to adjust the model to fit the Chemformer 2.0 code. I ran the following code to convert the model before using it.
chemUSPTO50 = torch.load('Chemformer USPTO-50k.ckpt')
chemUSPTO50['hyper_parameters']['vocabulary_size'] = chemUSPTO50['hyper_parameters'].pop('vocab_size')
torch.save(chemUSPTO50, 'Chemformer2 USPTO-50k.ckpt')If you know why this is happening, or if you are using a different model for the results below please let me know. Thank you.

This is the output on USPTO-15K dataset; a few questions on the output (I'm new to the pharma domain)
The columns of the output file:
| original_smiles | prediction_0 | log_likelihood_0 | prediction_1 | log_likelihood_1 | prediction_2 | log_likelihood_2 | prediction_3 | log_likelihood_3 | prediction_4 | log_likelihood_4 | prediction_5 | log_likelihood_5 | prediction_6 | log_likelihood_6 | prediction_7 | log_likelihood_7 | prediction_8 | log_likelihood_8 | prediction_9 | log_likelihood_9
When I pretrained the model from scratch, the traning process hangs during the first epoch. The GPU usage is stuck at 100%. The environment I used is as follows:
torch=1.8.0
pytorch_lightning=1.2.3
deepspeed=0.3.12
Is there anyone else encountered the same problem, and can give me some advice to tackle it? Thank you!
hi
I want to know how to get uspto_50.pickle from the origin uspto_50k.csv ?
Hi! I just wanted to ask if the retrosynthesis experiments include the reagents or not.
Thank you already!
I'm looking to run some predictions using the pretrained models you've uploaded (specifically models/fine-tuned/uspto-mixed/last.ckpt), and I've run into issues where pytorch lightning doesn't detect any hyperparams in the checkpoint model. I realize you've uploaded the hyperparams separately, but I'm not sure how to combine them into the model so that pytorch-lightning can load it correctly. Here is the exact error message I get:
>> $ python -m molbart.predict --reactants_path tests/test.csv --model_path uspto_mixed/last.ckpt --task forward_prediction
File "/baldig/chemistry/2023_rp/Chemformer/molbart/models/chemformer.py", line 387, in _initialize_from_ckpt
self.model_path, decode_sampler=self.sampler
File "/home/mcangel/anaconda3/envs/chemformer/lib/python3.7/site-packages/pytorch_lightning/core/saving.py", line 156, in load_from_checkpoint
model = cls._load_model_state(checkpoint, strict=strict, **kwargs)
File "/home/mcangel/anaconda3/envs/chemformer/lib/python3.7/site-packages/pytorch_lightning/core/saving.py", line 198, in _load_model_state
model = cls(**_cls_kwargs)
TypeError: __init__() missing 1 required positional argument: 'vocabulary_size'
If you have any ideas please let me know!
Dear Chemformer Team,
I cloned your repo and tried to set up the conda env with your env.yml:
conda env create -f env.yaml
However, that simply prompts a massive list of unfound packages and aborts. Is there an updated / more lenient version of the env.yaml or some container you could offer?
Thanks a lot - Tobi
Collecting package metadata (repodata.json): done
Solving environment: failed
ResolvePackageNotFound:
- libgomp==12.2.0=h65d4601_19
- pillow==9.2.0=py37h850a105_2
- pixman==0.40.0=h36c2ea0_0
- fontconfig==2.14.2=h14ed4e7_0
- mkl==2021.4.0=h06a4308_640
- libstdcxx-ng==12.2.0=h46fd767_19
- libuuid==2.32.1=h7f98852_1000
- pcre==8.45=h9c3ff4c_0
- intel-openmp==2021.4.0=h06a4308_3561
- xorg-libice==1.0.10=h7f98852_0
- numpy==1.21.5=py37h6c91a56_3
- libopenblas==0.3.21=pthreads_h78a6416_3
- _libgcc_mutex==0.1=conda_forge
- sqlite==3.40.0=h4ff8645_0
- python==3.7.11=h12debd9_0
- xorg-libxdmcp==1.1.3=h7f98852_0
- readline==8.2=h8228510_1
- libglib==2.68.4=h3e27bee_0
- libpng==1.6.39=h753d276_0
- libwebp-base==1.3.0=h0b41bf4_0
- lcms2==2.14=h6ed2654_0
- brotlipy==0.7.0=py37h27cfd23_1003
- boost-cpp==1.74.0=h312852a_4
- icu==68.2=h9c3ff4c_0
- certifi==2022.12.7=py37h06a4308_0
- libdeflate==1.14=h166bdaf_0
- xorg-renderproto==0.11.1=h7f98852_1002
- openjpeg==2.5.0=h7d73246_1
- libuv==1.44.2=h5eee18b_0
- xorg-kbproto==1.0.7=h7f98852_1002
- cairo==1.16.0=h6cf1ce9_1008
- expat==2.5.0=h27087fc_0
- pyopenssl==23.0.0=py37h06a4308_0
- xorg-xextproto==7.3.0=h0b41bf4_1003
- pysocks==1.7.1=py37_1
- freetype==2.12.1=hca18f0e_1
- urllib3==1.26.14=py37h06a4308_0
- lerc==4.0.0=h27087fc_0
- libgfortran5==12.2.0=h337968e_19
- libffi==3.3=h58526e2_2
- libsqlite==3.40.0=h753d276_0
- tk==8.6.12=h27826a3_0
- cffi==1.15.1=py37h74dc2b5_0
- xorg-libxrender==0.9.10=h7f98852_1003
- numpy-base==1.21.5=py37ha15fc14_3
- bzip2==1.0.8=h7f98852_4
- libiconv==1.17=h166bdaf_0
- libzlib==1.2.13=h166bdaf_4
- ncurses==6.3=h27087fc_1
- ca-certificates==2023.05.30=h06a4308_0
- mkl-service==2.4.0=py37h7f8727e_0
- cudatoolkit==11.1.74=h6bb024c_0
- xorg-libx11==1.8.4=h0b41bf4_0
- libxcb==1.13=h7f98852_1004
- xorg-libxext==1.3.4=h0b41bf4_2
- boost==1.74.0=py37h796e4cb_5
- libgcc-ng==12.2.0=h65d4601_19
- zlib==1.2.13=h166bdaf_4
- mkl_fft==1.3.1=py37hd3c417c_0
- xorg-libxau==1.0.9=h7f98852_0
- setuptools==59.8.0=py37h89c1867_1
- cryptography==39.0.1=py37h9ce1e76_0
- _openmp_mutex==4.5=2_gnu
- openssl==1.1.1u=h7f8727e_0
- xz==5.2.6=h166bdaf_0
- ninja==1.10.2=h06a4308_5
- pandas==1.3.5=py37he8f5f7f_0
- rdkit==2020.09.1=py37h0c252aa_0
- zstd==1.5.2=h3eb15da_6
- jpeg==9e=h0b41bf4_3
- ld_impl_linux-64==2.40=h41732ed_0
- gettext==0.21.1=h27087fc_0
- mkl_random==1.2.2=py37h51133e4_0
- libgfortran-ng==12.2.0=h69a702a_19
- idna==3.4=py37h06a4308_0
- pycairo==1.21.0=py37h0afab05_1
- pthread-stubs==0.4=h36c2ea0_1001
- requests==2.28.1=py37h06a4308_0
- xorg-libsm==1.2.3=hd9c2040_1000
- libgcc==7.2.0=h69d50b8_2
- ninja-base==1.10.2=hd09550d_5
- xorg-xproto==7.0.31=h7f98852_1007
- libtiff==4.4.0=h82bc61c_5
Hi @EBjerrum , I followed the steps in creating new venv and also downloaded the data/models from the link hosted by AZ.
I'm assuming I could try running predict.py and obtain the results connecting model path and the dataset path in the args.
However, I have a problem understanding what the args mean.
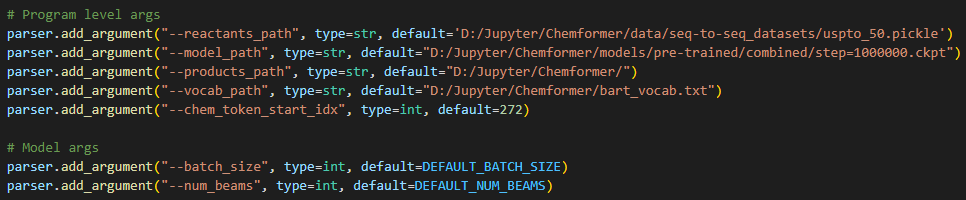
--reactants_path : Is this the pickle file dataset USPTO_50 for example? but the code I think expects a text file
--model_path : the path here I have is from the downloaded pretrained model 'combined' version
--products_path : the path where dataframe is saved on
--vocab_path : the path of bart_vocab
The rest of them, just using default values.
Would like to get this working to put up a working dashboard on HF Space :) Thanks!
Hello,
I have seen that you collaborated with Nvidia on the Megatron version of Chemformer, which can perform interpolation between two given molecules. How is this implemented?

The source link for this image is as follows. https://github.com/NVIDIA/cheminformatics/blob/master/tutorial/Tutorial.md
Hi there,
I found that you are using the second item in the sequence for sentence-level representation, as shown here. I'm wondering why not taking the first token (the CLS token)?
Hi,
The same regression fine-tuning of 3 tasks simulataneously but adding the other solubility datasets as well from here , but before I go ahead with it, I'm curious if a thought was given to this or were there any disadvantages to this? maybe because the dataset isnt completely reliable.
Thanks
Dear Chemformer Team,
I am currently embarking on a project aiming to perform regression analysis using biological activity data (specifically, pXC50 values) with the pretrained Chemformer model. The objective is to predict activity values based on SMILES strings.
In the process of setting up my environment and preparing for fine-tuning, I encountered a closed issue #13
and a fork of the repository, which provided clear examples and scripts for fine-tuning Chemformer on regression tasks. Notably, these resources referenced RegPropDataModule(_AbsDataModule) in finetune_regression_modules.py, suggesting it as a viable option for regression with Chemformer.
However, upon revisiting the Chemformer repository, it appears that the finetune_regression directory and RegPropDataModule class are no longer present in the example_scripts folder, which has left me uncertain about the best approach to undertake my regression task with the latest codebase.
With the above context, I am reaching out to seek your guidance on several points:
Current Recommended DataModule: Given the removal of RegPropDataModule and associated fine-tuning examples, could you advise on which DataModule in the current code structure is best suited for handling a dataset of SMILES strings with pXC50 values for regression analysis?
Script Selection: Among the scripts present in the repository (e.g., fine_tune.py, inference_score.py, predict.py), which would you recommend for fine-tuning the pretrained model on a regression dataset and for making subsequent predictions?
Further Recommendations: If there are any specific recommendations regarding data preprocessing, hyperparameter selection, or other considerations to optimize the use of Chemformer for this regression task, I would be grateful for your insights.
Thank you very much for your time and support !
Since we dont have access to the weights of the fine-tuned model for molecule property prediction task, I ask how do we fine-tune for this task using the Chemformer codebase?
I see for finetune we use: python -m molbart.example_scripts.finetune_regression.finetuneRegr.py <args>
Below are just the default args, but I'd like to know how would the data_path file look like for molecule property prediction task?
DEFAULT_data_path = 'all_TrainTestVal_pXC50.csv'
DEFAULT_vocab_path = "prop_bart_vocab.txt"
DEFAULT_study_name = "TL_Regr_pXC50_" + str(datetime.datetime.now())
DEFAULT_model_path = "pt_zinc_bart.ckpt"
DEFAULT_genes_path = "/individual_files"
DEFAULT_BATCH_SIZE = 32
DEFAULT_ACC_BATCHES = 1
DEFAULT_GRAD_CLIP = 1.0
DEFAULT_SCHEDULE = "cycle"
DEFAULT_AUGMENT = True
DEFAULT_WARM_UP_STEPS = 3000
DEFAULT_TRAIN_TOKENS = None
DEFAULT_NUM_BUCKETS = 24
DEFAULT_LIMIT_VAL_BATCHES = 1.0
DEFAULT_EPOCHS = 150
DEFAULT_GPUS = 1
DEFAULT_D_PREMODEL = 512
DEFAULT_MAX_SEQ_LEN = 300
DEFAULT_LR = 3e-4
DEFAULT_H_FEEDFORWARD = 2048
DEFAULT_drp = 0.2
DEFAULT_Hdrp = 0.4
DEFAULT_WEIGHT_DECAY = 0.0
I have the 3 datasets with me: FreeSolv, Lipophilicity and ESOL.
Thanks in advance !
Not an issue with the code but just letting you know that the pdf link at the accepted Chemformer version in Mach Learn: Sci Technol doesn't actually link to your article. It has your title on the front page but the rest of the pdf is an article called "Physics makes the difference: Bayesian optimization and active learning via augmented Gaussian process" by Ziatdinov et al. Just in case you want to tell the journal to correct it.
A declarative, efficient, and flexible JavaScript library for building user interfaces.
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
An Open Source Machine Learning Framework for Everyone
The Web framework for perfectionists with deadlines.
A PHP framework for web artisans
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
Some thing interesting about web. New door for the world.
A server is a program made to process requests and deliver data to clients.
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
Some thing interesting about visualization, use data art
Some thing interesting about game, make everyone happy.
We are working to build community through open source technology. NB: members must have two-factor auth.
Open source projects and samples from Microsoft.
Google ❤️ Open Source for everyone.
Alibaba Open Source for everyone
Data-Driven Documents codes.
China tencent open source team.