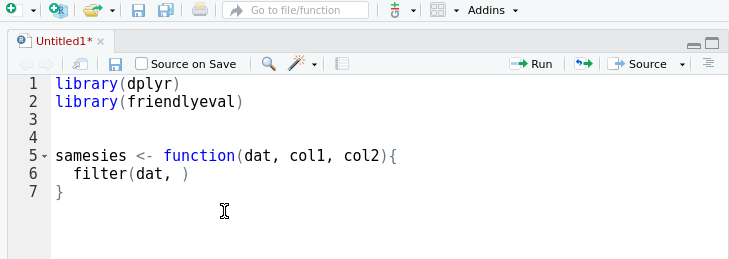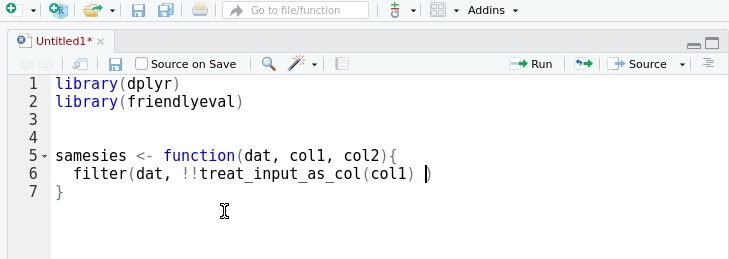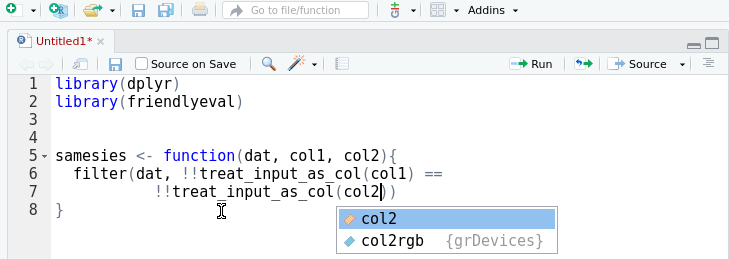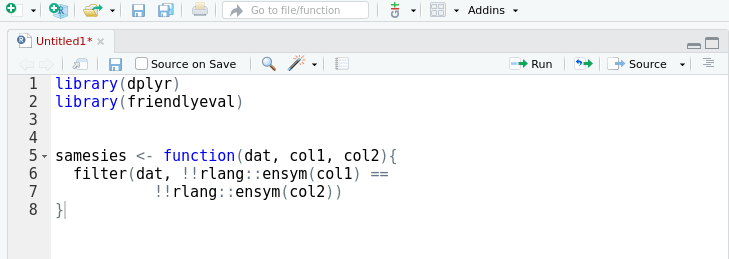
A friendly interface to the tidy eval framework and the
rlang package for casual
dplyr users.
This package provides an alternative, auto-complete friendly interface to
rlang that is more closely aligned with the task domain of a user 'programming
with dplyr'. It implements most of the cases in the 'programming with
dplyr' vignette.
The interface can also convert itself to standard rlang with the help of an
RStudio addin that replaces
friendlyeval functions with their rlang equivalents. This allows you to
prototype in friendly, then subsequently automagically transform to rlang.
Your friends won't know the difference.
devtools::install_github("milesmcbain/friendlyeval")Arguments passed to dplyr can be treated as:
- a single literal column name (e.g.
mpginselect(mtcars, mpg)) - a single expression (e.g.
cyl <= 6infilter(mtcars, cyl <= 6)) - a list of literal column names (e.g.
mpg, cylinselect(mtcars, mpg, cyl)) - a list of expressions (e.g.
hp >= mean(hp), wt > 3infilter(mtcars, hp >= mean(hp), wt > 3))
dplyr uses special argument handling to interpret and treat those arguments as
one or more column names or expressions. User functions don't perform that same
argument handling, so we need some way to tell dplyr how to treat these
arguments we pass from our enclosing functions. rlang provides the functions
we need to do just that, but knowing which rlang function maps to each use
case requires a fairly nuanced understanding of metaprogramming concepts.
friendlyeval helps bridge that gap by providing a descriptive (and
auto-complete friendly) set of eight complimentary functions that instruct
dplyr to resolve arguments we pass using either:
- the literal input provided as the arguments to our function (e.g. the text
latandloninmy_select(dat, lat, lon)) - the string values of those arguments (e.g.
"lat"and"lon"inmy_select(dat, arg1 = "lat", "lon")):
| function | usage |
|---|---|
treat_input_as_col |
Treat the literal text input provided as a dplyr column name. |
treat_input_as_expr |
Treat the literal text input provided as an expression involving a dplyr column name (e.g. in filter(dat, col1 == 0), col1 == 0 is an expression involving the value of col1). |
treat_inputs_as_cols |
Treat a comma separated list of arguments as a list of dplyr column names. |
treat_inputs_as_exprs |
Treat a comma separated list of arguments as a list of expressions. |
treat_string_as_col |
Treat the character value of your function argument as a dplyr column name. |
treat_string_as_expr |
Treat the character value of your function argument as an expression involving a dplyr column name. |
treat_strings_as_cols |
Treat a list of character values as a list of dplyr column names. |
treat_strings_as_exprs |
Treat a vector of strings as a list of expressions involving dplyr column names. |
These eight functions are used in conjunction with three tidy eval operators:
!!!!!:=
!! and !!! are signposts that tell dplyr:
"Stop! This needs to be evaluated to resolve column names or expressions".
!! tells dplyr to expect a single column name or expression, whereas !!! says to expect a list of column names or expressions.
:= is used in place of = in the special case where we need dplyr to
resolve a column name on the left hand side of an = like in
mutate(!!treat_input_as_col(colname) = rownumber). Evaluating on the left hand
side in this example is not legal R syntax, so instead we must write:
mutate(!!treat_input_as_col(colname) := rownumber).
dplyr functions try to be user-friendly by saving you typing. This allows you
to write code like mutate(data, col1 = abs(col2), col3 = col4*100) instead of
the more cumbersome base R style: data$col <- abs(data$col2); data$col3 <- data$col4*100.
The cost of this convenience is more work when we want to write functions that call dplyr, because dplyr needs to be instructed how to treat the arguments we pass to it. For example, this function does not work as we might expect:
double_col <- function(dat, arg){
mutate(dat, result = arg*2)
}
double_col(mtcars, cyl)
# Error in mutate_impl(.data, dots) :
# Evaluation error: object 'cyl' not found.This is because our double_col function doesn't perform the same special argument handling as dplyr functions. What if we pass our column name as a string value instead?
double_col(mtcars, arg = 'cyl')
# Error in mutate_impl(.data, dots) :
# Evaluation error: non-numeric argument to binary operator.That doesn't work either, even though those were our only options under normal evaluation rules! Fortunately, there are two ways to make double_col work. We can either:
- Instruct
dplyrto treat the literal input provided for theargargument as a column name. Sodouble_col(mtcars, cyl)would work. - Instruct
dplyrto treat the string value bound toarg- "cyl" - as a column name, rather than as a normal character vector. Sodouble_col(mtcars, arg = "cyl")would work.
Using what was input, dplyr style:
double_col <- function(dat, arg){
mutate(dat, result = !!treat_input_as_col(arg) * 2)
}
## working call form:
double_col(mtcars, cyl)Using the supplied value:
double_col <- function(dat, arg){
mutate(dat, result = !!treat_string_as_col(arg) * 2)
}
## working call form:
double_col(mtcars, arg = 'cyl')A more useful version of double_col would be to allow the name of the resulting column to be set via the function. Again, this can be done using the literal input, dplyr style:
double_col <- function(dat, arg, result){
## note usage of ':=' for lhs eval.
mutate(dat, !!treat_input_as_col(result) := !!treat_input_as_col(arg) * 2)
}
## working call form:
double_col(mtcars, cyl, cylx2) Or using supplied values:
double_col <- function(dat, arg, result){
## note usage of ':=' for lhs eval.
mutate(dat, !!treat_string_as_col(result) := !!treat_string_as_col(arg) * 2)
}
## working call form:
double_col(mtcars, arg = 'cyl', result = 'cylx2')When wrapping group_by, you will likely want to pass a list of column names. Here's how to do that using what was input, dplyr style:
reverse_group_by <- function(dat, ...){
## this expression is split out for readability, but it can be nested into below.
groups <- treat_inputs_as_cols(...)
group_by(dat, !!!rev(groups))
}
## working call form
reverse_group_by(mtcars, gear, am)Here's how to do it using a list of values:
reverse_group_by <- function(dat, columns){
groups <- treat_strings_as_cols(columns)
group_by(dat, !!!rev(groups))
}
## working call form:
reverse_group_by(mtcars, c('gear', 'am'))And here's how to do it using the values of ...:
reverse_group_by <- function(dat, ...){
## note the list() around ... to collect the values of the arguments into a list.
groups <- treat_strings_as_cols(list(...))
group_by(dat, !!!rev(groups))
}
## working call form:
reverse_group_by(mtcars, 'gear', 'am')Using the _expr functions, you can pass expressions involving column names to
dplyr functions like filter, mutate and summarise. An example of a
function involving an expression is a more general version of the double_col
function from above, called double_anything, that can take expressions
involving columns, and double those:
double_anything <- function(dat, arg){
mutate(dat, result = (!!treat_input_as_expr(arg)) * 2)
}
## working call form:
double_anything(mtcars, cyl * am)
## mpg cyl disp hp drat wt qsec vs am gear carb result
## 1 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4 12
## 2 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4 12
## 3 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1 8
## 4 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1 0
## 5 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2 0A common usage pattern is to take a list of expressions. Consider the filter_loudly function that reports the number of rows filtered:
filter_loudly(mtcars, cyl >= 6, am == 1)
## Filtered out 27 rows.
## mpg cyl disp hp drat wt qsec vs am gear carb
## 1 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
## 2 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
## 3 15.8 8 351 264 4.22 3.170 14.50 0 1 5 4
## 4 19.7 6 145 175 3.62 2.770 15.50 0 1 5 6
## 5 15.0 8 301 335 3.54 3.570 14.60 0 1 5 8 You can implement this function using filtering expressions exactly as input, dplyr style:
filter_loudly <- function(x, ...){
in_rows <- nrow(x)
out <- filter(x, !!!treat_inputs_as_exprs(...))
out_rows <- nrow(out)
message("Filtered out ",in_rows-out_rows," rows.")
return(out)
}
## working call form:
## filter_loudly(mtcars, cyl >= 6, am == 1) Or using a list/vector of values, parsed as expressions:
filter_loudly <- function(x, filter_expressions){
## if accepting list arguments, should check all are character
stopifnot(purrr::every(filter_expressions, is.character))
in_rows <- nrow(x)
out <- filter(x, !!!treat_strings_as_exprs(filter_expressions))
out_rows <- nrow(out)
message("Filtered out ",in_rows-out_rows," rows.")
return(out)
}
## working call form:
## filter_loudly(mtcars, list('cyl >= 6', 1))Or capturing the values of ..., parsed as expressions:
filter_loudly <- function(x, ...){
dots <- list(...)
## if accepting list arguments, should check all are character
stopifnot(purrr::every(dots, is.character))
in_rows <- nrow(x)
out <- filter(x, !!!treat_strings_as_exprs(dots))
out_rows <- nrow(out)
message("Filtered out ",in_rows-out_rows," rows.")
return(out)
}
## working call form:
## filter_loudly(mtcars, 'cyl >= 6', 'am == 1')It may have occurred to you that there are cases where a column name is a valid expression and vice versa. This is true, and it means that in some situations, you could switch the _col and the _expr versions of functions (e.g. use treat_input_as_expr in place of treat_input_as_col) and things would continue to work. However, using the col version where appropriate invokes checks that assert that what was passed to it can be interpreted as a simple column name. This is useful in situations where expressions are not permitted, like in select or on the left hand side of the internal assignment in mutate: e.g. mutate(lhs_col = some_expr).