A platform for advanced Machine Learning research and applications.
The goal of rtemis is to make data science efficient and accessible with no compromise on flexibility.
R version 4.1 or higher
You can install rtemis from r-universe or using pak, remotes, or devtools.
-
r-universe:install.packages('rtemis', repos = c('https://egenn.r-universe.dev', 'https://cloud.r-project.org'))
-
pak:pak::pkg_install("egenn/rtemis")
-
remotes:remotes::install_github("egenn/rtemis")
-
devtools:devtools::install_github("egenn/rtemis")
To allow compilation from source of any dependencies that require Fortran, you will need to install the GNU Fortran compiler. The easiest way to do this is with Homebrew:
brew install gccThen, you will need to add the following to your ~/.R/Makevars file:
FC = usr/local/opt/gcc/bin/gfortran
F77 = /usr/local/opt/gcc/bin/gfortran
FLIBS = -L/usr/local/opt/gcc/libd_UMAP() requires the uwot package, which currently requires that the Matrix and
irlba dependencies be installed from source. See more in the uwot issue
here.
See here for more setup and installation instructions.
Note: Make sure to keep your installation updated by running
remotes::install_github("egenn/rtemis") regularly: it will only proceed if
there are updates available.
Install dependencies if they are not already installed:
packages <- c("future.apply", "ranger")
.add <- !packages %in% installed.packages()
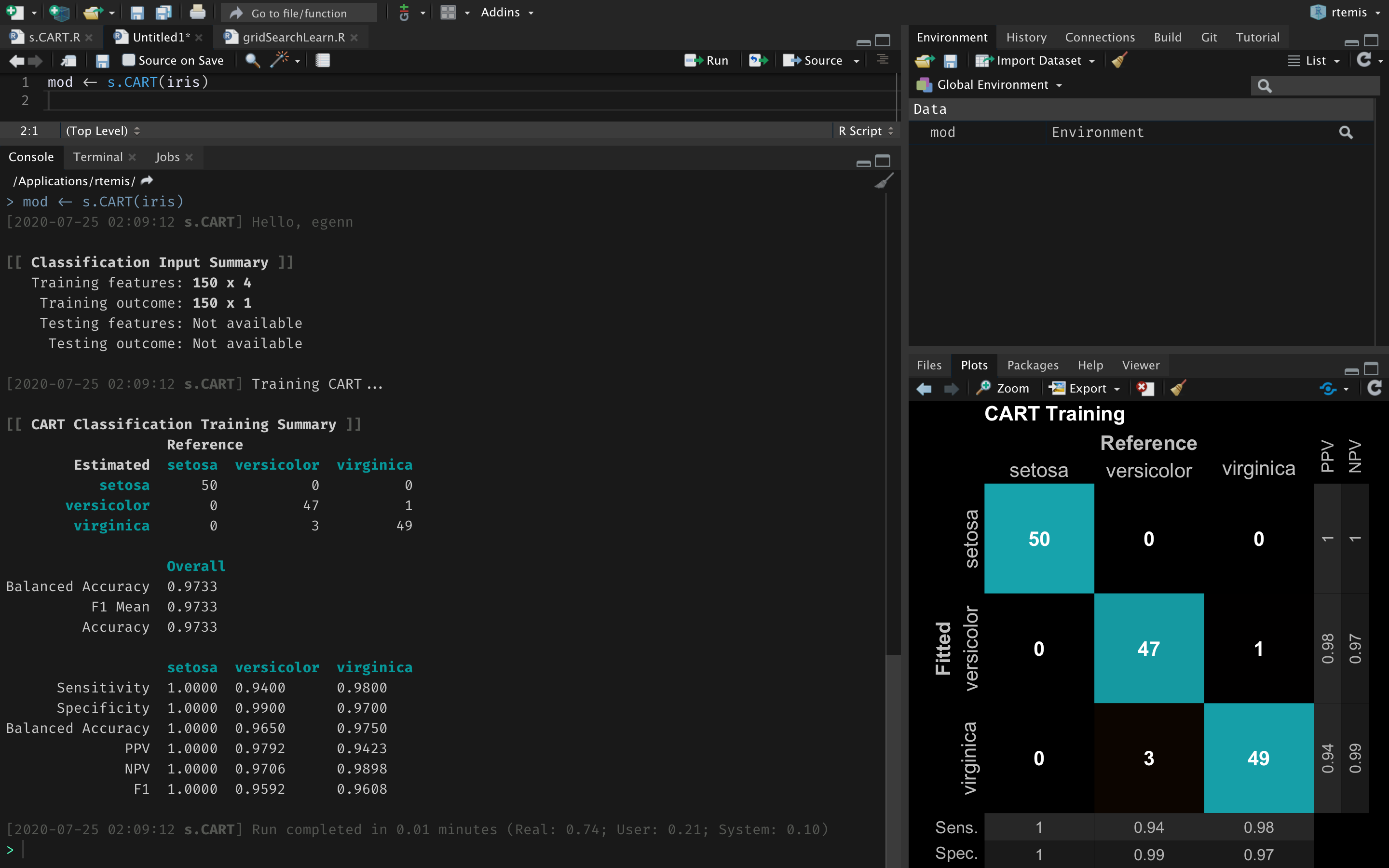
install.packages(packages[.add])Get cross-validated random forest performance on the iris dataset:
library(rtemis)
mod <- train_cv(iris)We are working towards the 1.0 release, which will feature updates to the
API as well as the backend, and preparing for CRAN submission.
This will be accompanied by expansion of the documentation.
For all updates, please see the NEWS file.
The Python and Julia ports, rtemispy and Rtemis.jl, which are not yet
publicly available, are in parallel development. With the upcoming 1.0 release
of rtemis, the aim is to offer a unified API across all three languages.
-
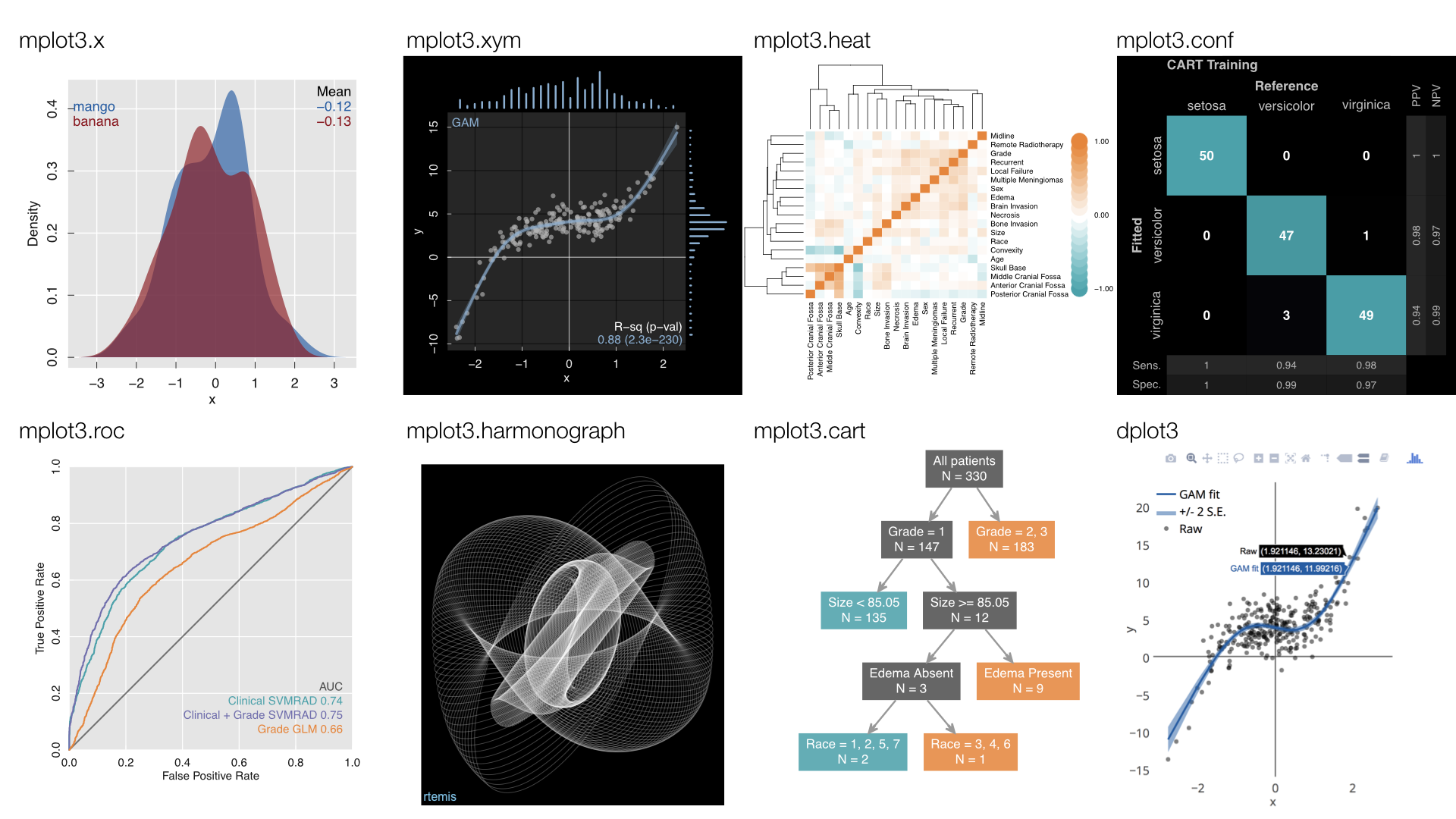
Visualization
- Static: mplot3 family (base graphics)
- Dynamic: dplot3 family (plotly)
-
Unsupervised Learning
- Clustering: c*_
- Decomposition: d*_
-
Supervised Learning
- Classification, Regression, Survival Analysis: s*_
-
Cross-Decomposition
- Sparse Canonical Correlation / Sparse Decomposition: x*_
-
Meta-Models
- Model Stacking: metaMod()
- Modality Stacking: metaFeat()
- Group-weighted Stacking: metaGroup()
(metaFeat and metaGroup have been removed for updating)
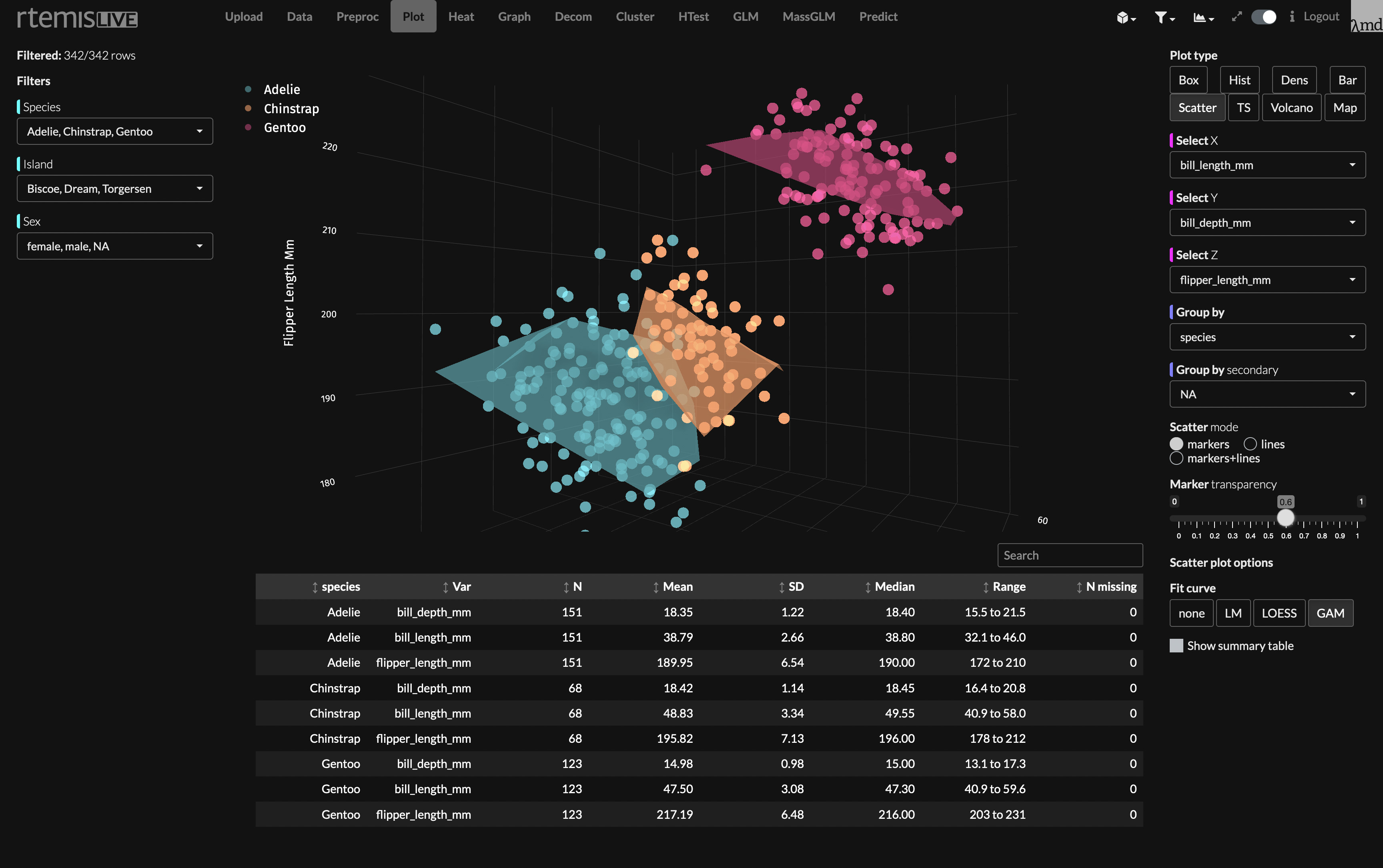
rtemislive is rtemis' web interface / GUI. It makes advanced visualization and modeling instantly accessible by all. It is currently available for beta testing at UCSF, and will be made publicly available once funding is secured for a hosting server.
Get the rtemis-dark VS Code theme.
Recommended font is Fira Code with its pretty ligatures.
Python and Julia APIs are in development. The goal is to delliver a unified API across all three languages by the time of the 1.0 release.